Systems biology relies heavily on modeling technologies to map out the various networks and pathways that exist in systems under study. Researchers use a variety of tools to make dynamic models that can be used to visualize information or to collect data by creating a virtual system.
Cytoscape:
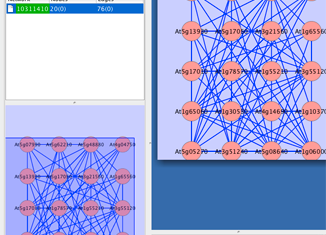 Cytoscape is a powerful modeling software employed by systems biology researchers to create network diagrams. Its simple, Java-based interface allows the user to create systems with various nodes and edges to model the components and interactions that exist within a network. The user begins by creating a spreadsheet listing all of the nodes and edges, mapping out the interactions that occur in the system, which is then imported into the program and converted into a diagram. Cytoscape provides an elegant method of visualizing systems information and is a widely-used software among researchers today.
Cytoscape is a powerful modeling software employed by systems biology researchers to create network diagrams. Its simple, Java-based interface allows the user to create systems with various nodes and edges to model the components and interactions that exist within a network. The user begins by creating a spreadsheet listing all of the nodes and edges, mapping out the interactions that occur in the system, which is then imported into the program and converted into a diagram. Cytoscape provides an elegant method of visualizing systems information and is a widely-used software among researchers today.
The program can also be used in advanced research with the use of data sets. Cytoscape allows the user to input data along with nodes, define equations within the edges, and run controlled experiments, considering the factors of perterbations, in order to gain real-world data that is manifested in the model.
Stella:
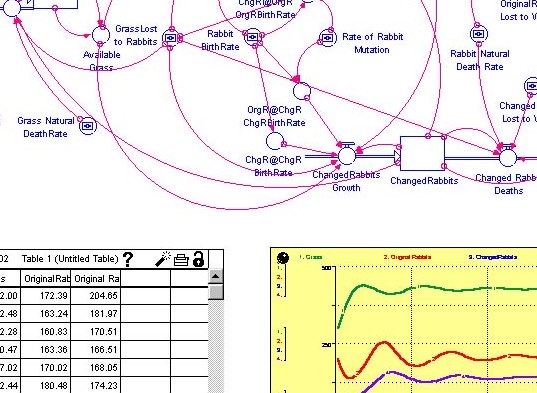 Stella is a modeling software, similar to Cytoscape, which is used to create network diagrams and system models. Stella, however, has a simpler interface and is less prominant as a research tool. It emphasizes the process of building models and learning about systems rather than research, as it was designed as an education software.
Stella is a modeling software, similar to Cytoscape, which is used to create network diagrams and system models. Stella, however, has a simpler interface and is less prominant as a research tool. It emphasizes the process of building models and learning about systems rather than research, as it was designed as an education software.
Students can use Stella to create their own networks by adding elements and defining the rates at which the elements change, so its common applications include food webs and population growth studies. The conditions created can be modeled to provide usable data about the growth or decline of the system. Data can also be entered into the program to recreate the system that yielded a set of data, so Stella is a great analysis tool for real-world data and publications.
