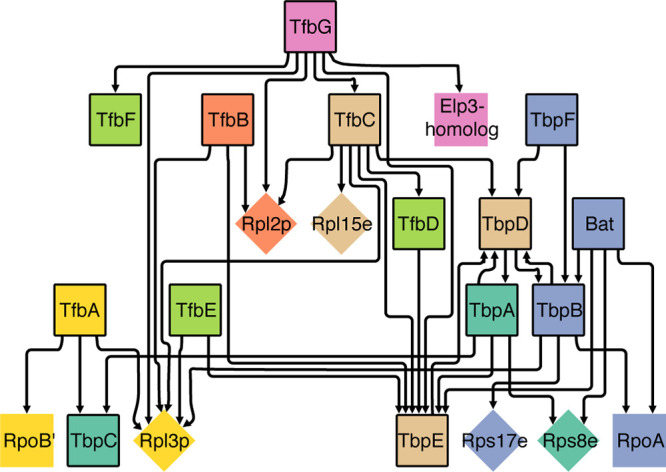
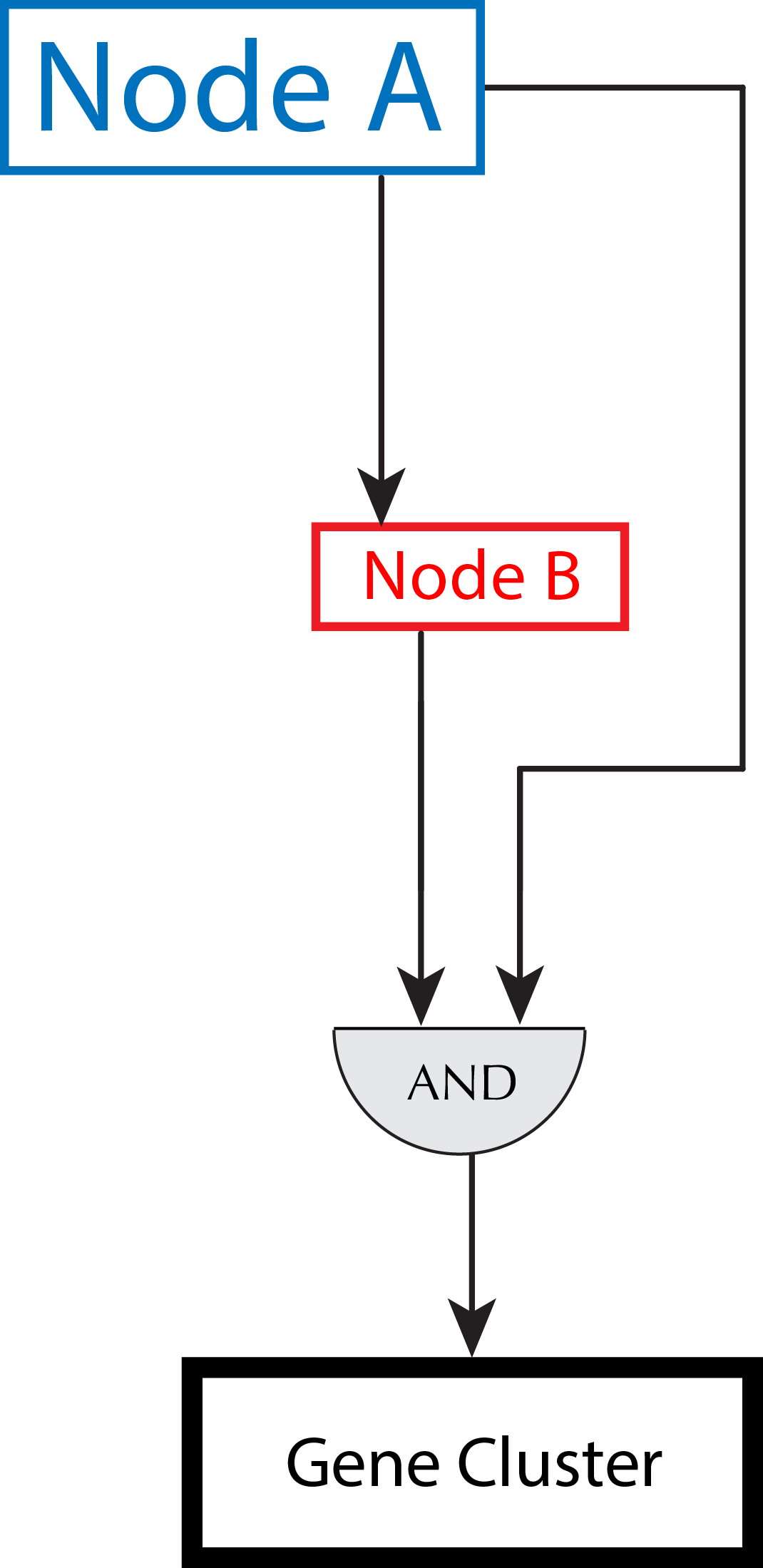
Summary: In this project we seek to investigate how modularity of transcriptional programs generated through expansions of general transcription factors (GTFs) begets the extraordinary success of microbes in adapting to diverse niches. Duplications or even small changes to the regulation or interactions of a GTF dramatically alters the modularity of a transcriptional regulatory network. We have discovered that modular […]
Read moreArchives: Projects
A post type for your projects
Project Template
Development of multicellular organisms from a single cell is arguably one of the most fascinating phenomena in biology. Mechanistic understanding of such a complex process is an important scientific challenge. Our single cell work aligns with the development of a systems perspective framework to comprehend how individual cellular states emerge and collectively result in a homeostatic multicellular organism.
Read moreEGRIN
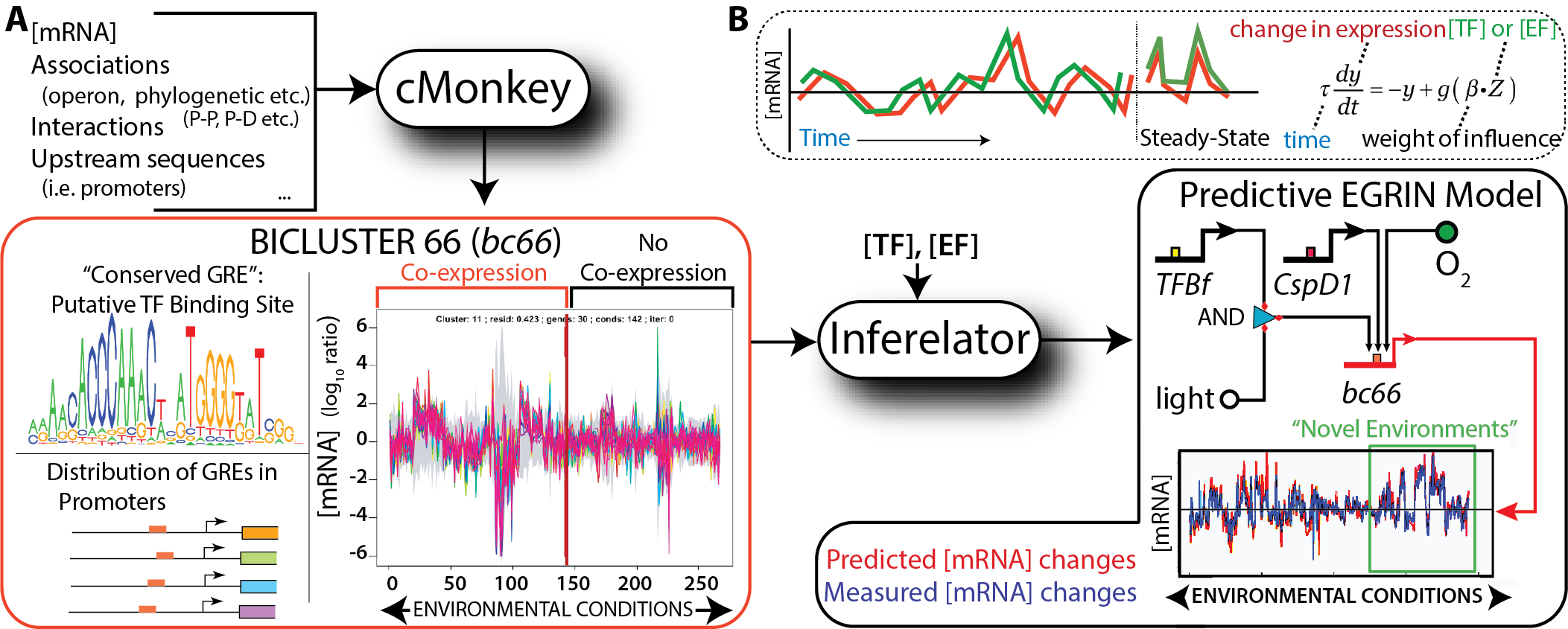
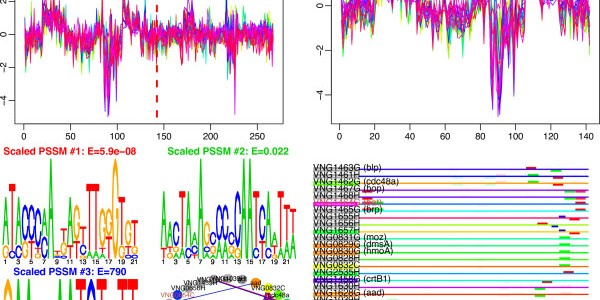
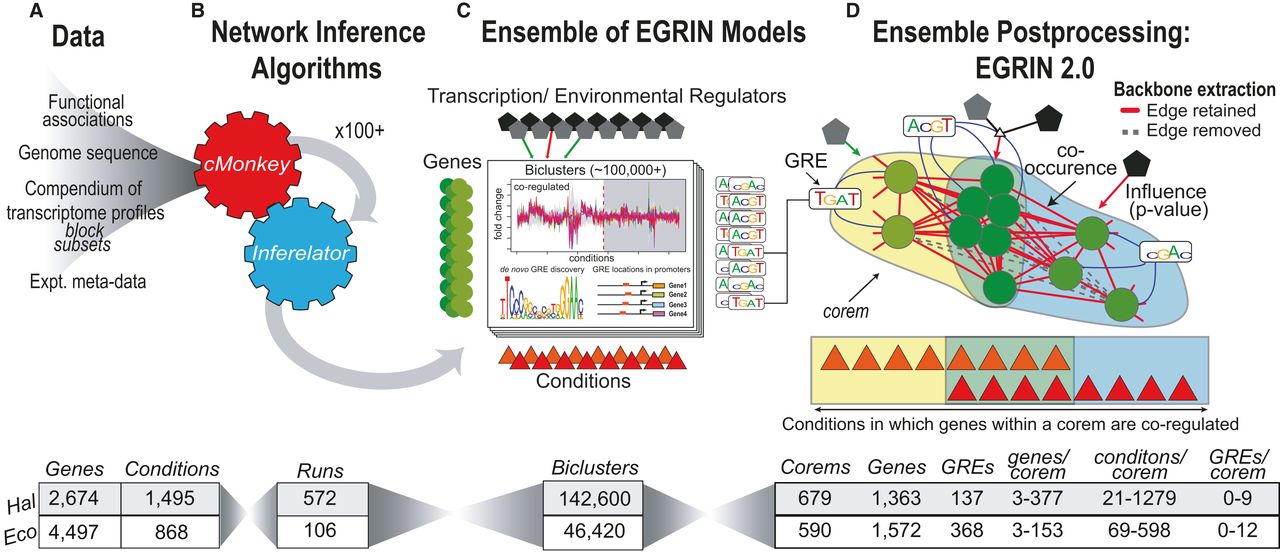
EGRIN (Environment and Gene Regulatory Influence Network). We have developed an approach for data-driven inference of a systems-scale predictive GRN model for any organism that can be cultured in the laboratory. The computational framework is comprised of two algorithms: cMonkey for data integration and biclustering, and Inferelator for inference of regulatory influences. Briefly, cMonkey integrates gene expression, GRE analysis, and […]
Read morecMonkey2
cMonkey2 – Python port of the cMonkey biclustering algorithm Description This is the Python implementation of the cMonkey algorithm based on the original R implementation by David J. Reiss, Institute for Systems Biology. Documentation A complete set of documentation for installation and running of cMonkey is on the project’s Github Pages. There are also developer and user discussion groups. Contact Please report all bugs or other issues […]
Read morecMonkey
You have reached the updated cMonkey site with a new R package, installation and running instructions, and additional information. If you are looking for the original cMonkey site, you can find it here. The original cMonkey algorithm was published with the manuscript “Integrated biclustering of heterogeneous genome-wide datasets for the inference of global regulatory networks”, by David J Reiss, Nitin S Baliga and Richard Bonneau. *NEW* cMonkey package and source code are now on […]
Read moreHalobacterium species NRC-1 Genome
Search SBEAMS Database Select attributes All AttributesGene SymbolORF NameFull Gene NameEC Number Enter Keywords Action Search Show all domain search results in SBEAMS for Halobacterium sp. NRC-1 (new window) Explore data with the Halobacterium Gaggle (new window) Data Download Loading….
Read moreSystems Education Experiences
The Systems Education Experiences (SEE) program provides an integrative approach to help all participants internalize systems biology methods and thinking. Our Vision is for all students to be engaged in using systems-level, scientific thinking to better understand and participate in solving today’s complex problems. Our Purpose is to cultivate cross-disciplinary skills for solving complex problems. We do this through an […]
Read moreProjectFeed1010
A human crisis of unprecedented proportions is in the making. Diminishing arable land, water shortage, and expected population growth to 10 billion portend a catastrophic food security crisis by 2030. This threat requires a dramatic reduction of the environmental footprint of agricultural practices. A paradigm shift to the food supply chain is necessary to meet an expected 70% or more […]
Read moreSingle Cell Network Inference
Development of multicellular organisms from a single cell is arguably one of the most fascinating phenomena in biology. Mechanistic understanding of such a complex process is an important scientific challenge. Our single cell work aligns with the development of a systems perspective framework to comprehend how individual cellular states emerge and collectively result in a homeostatic multicellular organism.
Read moreEGRIN2.0
We previously constructed an “Environment and Gene Regulatory Influence Network” (EGRIN) for Halobacterium salinarum NRC‐1 (Bonneau et al, 2007). This model was constructed in two steps. First, modular organization of gene regulation was deciphered through semi‐supervised biclustering of gene expression, guided by biologically informative priors and de novo cis‐regulatory GRE detection for module assignment (cMonkey; Reiss et al, 2006). Second, using a […]
Read moreGenetic Programs Controlling Responses of MTB
When MTB infects a human host, a menagerie of immune cells swarm around the pathogens and restrict their access to oxygen and nutrients, whereafter the bacteria cease to replicate and slow their metabolism to a crawl, entering into a latent state where they become particularly resistant to antibiotics. If, however, the host’s immune system is compromised, the bacteria once again […]
Read moreNetwork Portal
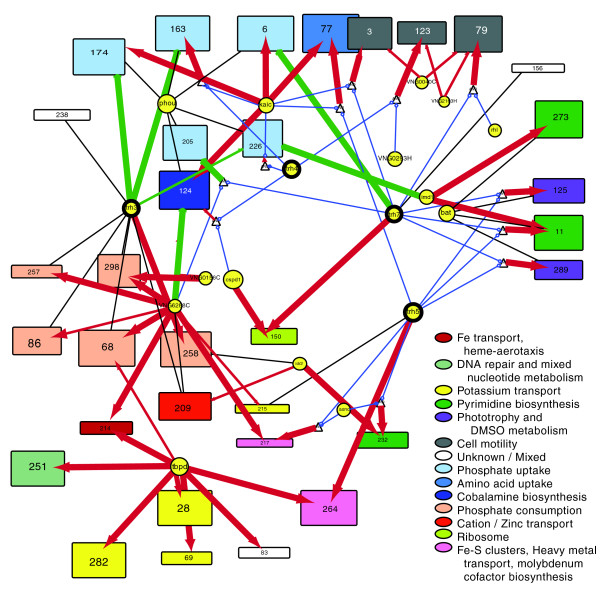
The Network Portal is a database of gene transcription regulatory networks and enables exploration, annotation and comparative analysis.Deciphering the complexity of biological systems requires a systems-level view of regulatory players and their interactions. However, inference of these complex interactions is challenging, and further, visualizing and analyzing these interactions often requires advanced computational expertise, tools and resources.Network Portal provides analysis and […]
Read more