 When MTB infects a human host, a menagerie of immune cells swarm around the pathogens and restrict their access to oxygen and nutrients, whereafter the bacteria cease to replicate and slow their metabolism to a crawl, entering into a latent state where they become particularly resistant to antibiotics. If, however, the host’s immune system is compromised, the bacteria once again become active, resuming their destruction within and in many cases killing the host. We use oxygen deprivation to induce a dormant state in the bacteria in vitro, to help mimic what happens when MTB infects a host. By pushing the experimental methods typically used to study this system, and elucidating the complex genetic circuits that might drive this behavior, We hope to shed light on the biology of latency in MTB, constructing predictive models of its genetic control systems and suggesting new routes by which to discover drugs to treat it.
When MTB infects a human host, a menagerie of immune cells swarm around the pathogens and restrict their access to oxygen and nutrients, whereafter the bacteria cease to replicate and slow their metabolism to a crawl, entering into a latent state where they become particularly resistant to antibiotics. If, however, the host’s immune system is compromised, the bacteria once again become active, resuming their destruction within and in many cases killing the host. We use oxygen deprivation to induce a dormant state in the bacteria in vitro, to help mimic what happens when MTB infects a host. By pushing the experimental methods typically used to study this system, and elucidating the complex genetic circuits that might drive this behavior, We hope to shed light on the biology of latency in MTB, constructing predictive models of its genetic control systems and suggesting new routes by which to discover drugs to treat it.
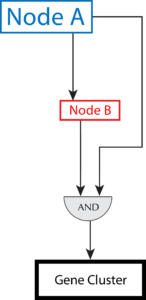
Transcription factors (TFs) modulate the expression of their downstream target genes through the simple functions of activation or repression. Over time, evolution has joined these simple functions into new emergent behaviors by linking together TFs into sub-circuits known as network motifs. We started by mining our existing Mycobacterium tuberculosis network for double-negative-feedback-loops. We then determined that a high-resolution time series experimental design would be better suited for observing network motifs in action. Therefore, we have shifted our efforts to discover network motifs that control the transition into a hypoxia-induced dormant phenotype through high resolution sampling of the transcriptomes of Mtb cells undergoing controlled hypoxia and subsequent reaeration. To conduct these experiments we have designed and developed an experimental setup that allows us to control the oxygen content and continuously monitor dissolved oxygen levels. These experiments will provide us with the required expression measurements to determine the validity of our network motif structure, and also provide the means to experimentally test specific genetic interventions that perturb these network motifs.
