Elisa Gehrke and Iman Janoo
—————————————–Abstract/Background—————————————–
Liquid chromatography-mass spectrometry (LC-MS)-based proteomics technologies, especially targeted MS e.g. Selected Reaction Monitoring (SRM), have been widely used in protein biomarker discovery for many different diseases. Due to the extremely wide dynamic range of the serum/plasma proteome (~10e11), it is still challenging to detect and measure serum proteins and peptides in very low abundance (for example, <1 µg/mL), without extensive fractionation or enrichment. The high amount of serum lipids and other types of micro and macro molecules also impact MS performance and assay consistency.
The Suspension Trap (S-Trap) digestion procedure developed by the biotechnology company ProtiFi, provides an alternative approach in serum sample preparation, with several advantages over the traditional in-solution digestion method, and with improved efficiency and yield of trypsin digestion.
We have recently modified the standard protocol of the S-Trap for use with depleted serum samples. In this study, to determine the more favorable method, S-Trap digestion was compared to traditional in-solution digestion in terms of peptide recovery.
————————————————-Objectives————————————————–
• Use blood samples and proteomic analyses to find peptide differences in patients considered obese and those considered healthy
• Use evidence to discover potential biomarkers for obesity
• Compare the effectiveness of traditional In-Solution method to Suspension Trap Digestion in protein samples
• Use mass spectrometry to analyze results of In-Solution and S-Trap runs
• Use mass spectrometry to determine which protein peptides are related to obesity
—————————————-What is a Biomarker?—————————————–
A biomarker is anything that can be measured that is indicative of a phenomenon such as a disease or an infection. The goal of this project is to find a biomarker that indicates a heightened risk for obesity. In order to achieve this, organ-specific, obesity-related proteins must be carefully chosen from a set of 300 before running them through the Mass Spectrometer and analyzing a full set.
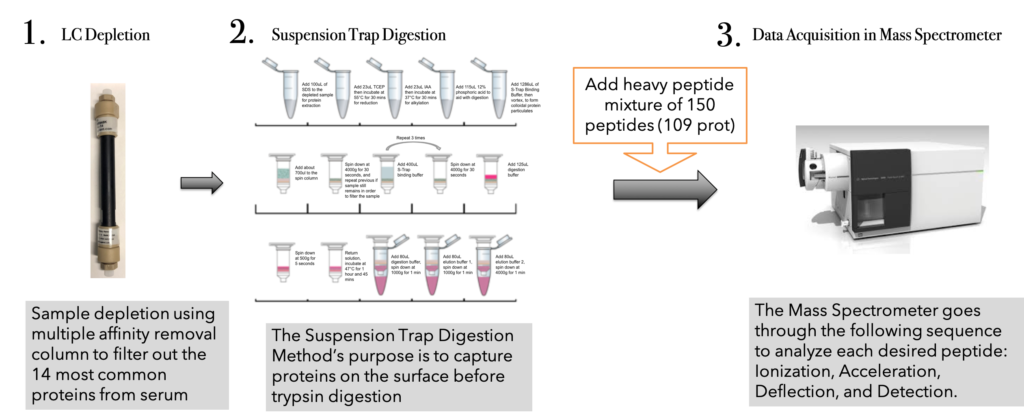
—————————————————-Design—————————————————–
——————Data: Evaluating S-Trap and In-Solution Digestion——————
For each individual peptide, we viewed the S-Trap and In-Solution graphs and decided which one had a higher intensity, and better transition curves. Based on these categories, we labeled the peptide with the method that was more effective.
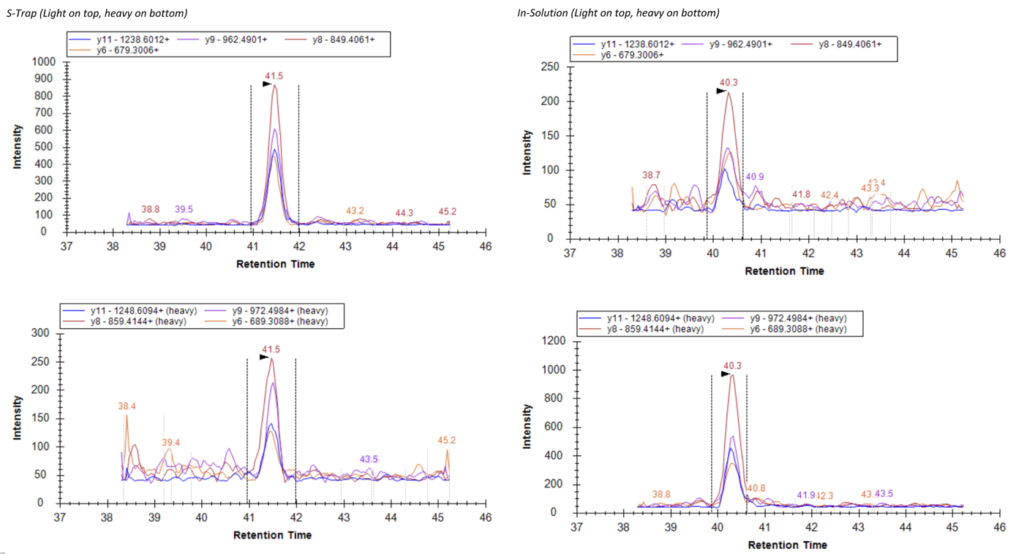
This figure shows the intensity for the light peptide is in the 900s between the retention times of 41 and 42 minutes. In contrast, the In-Solution method shows that the light peptide, between the same retention times, only had an intensity between 200 and 250. Additionally, the ratio of light to heavy peptide intensity for the S-Trap Digestion is 4:1, while the ratio of light to heavy peptide intensity for the In-Solution Digestion is 1:4.3. According to this example peptide, these methods are inverses of each other, and the ratio of the S-Trap was more desirable.
————————————————Conclusion————————————————–
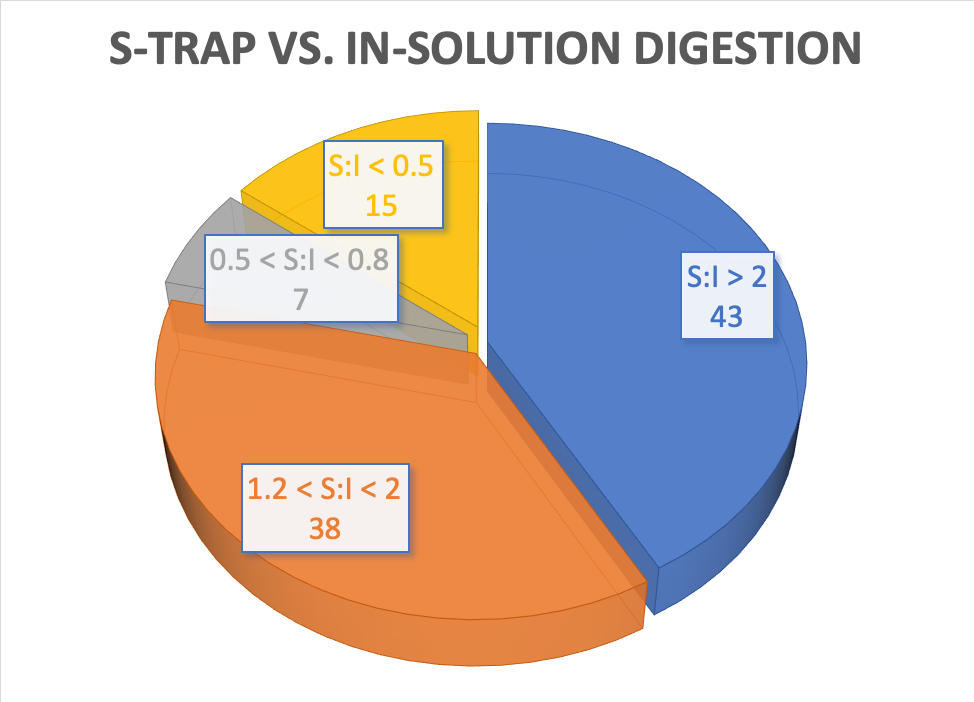
•Suspension Trap Digestion was generally more effective than In-Solution Digestion
•Suspension Trap Digestion is favorable to hydrophobic peptide. Most peptides performed better in the In-solution protocol are highly hydrophilic and in low retention time.
•With the more aggressive washes with SDS, phosphoric acid and methanol, suspension Trap Digestion efficiently removes lipids in serum, which produces much cleaner peptide products for mass spec analysis.
———————————————-Further Study———————————————–
•Further compare S-Trap and In-Solution digestion protocols in hydrophobicity, sensitivity, reproducibility, intra- and inter-batch variabilities
•Apply the modified S-Trap protocol in real clinical samples with obesity and Lyme disease to further investigate its effectiveness and limitations.
——————————————-Acknowledgements——————————————
Special thanks to Li Tang, Yong Zhou, Nathan Price, Leroy Hood, Claudia Ludwig, Rachel Calder, and Becky Howsmon for all of the mentorship and support throughout our seven weeks at ISB.