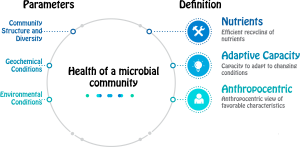
 Microbial communities influence the cycling of key nutrients including carbon, nitrogen, and sulfur. These cycles are tightly linked to the health of the planet, from greenhouse gas emissions to soil fertility. As anthropogenic disturbance continues to modify the natural environment, quantitative tools are critically needed in order to characterize the current state and predict the future health of these complex communities. In order to meet these demands, we are developing quantitative metrics of ‘health’ of a microbial community. We would like to understand, both biotic and abiotic factors, that are predictive of current and future health of a complex microbial community. Additionally, we aim to decipher the relationship between biodiversity, resistance (ability to maintain functional activity state in face of stress), resilience (ability to recover functional activity state after stressful perturbation), and adaptive capacity (ability to adopt new functional activity states in a new environment) of a microbial community? We pursue these studies by using microbial communities from Oak Ridge Environmental Field Research site and Fluidized Bed Reactors or FBRs. FBRs allow retention of attached populations (thereby retaining non-growth populations and system adaptive capacity, provide non-destructive sampling and can mimic rapid geochemical transitions (e.g. providing for short or prolonged periods of stress). Therefore, FBRs provide excellent system to study health of microbial communities.
Microbial communities influence the cycling of key nutrients including carbon, nitrogen, and sulfur. These cycles are tightly linked to the health of the planet, from greenhouse gas emissions to soil fertility. As anthropogenic disturbance continues to modify the natural environment, quantitative tools are critically needed in order to characterize the current state and predict the future health of these complex communities. In order to meet these demands, we are developing quantitative metrics of ‘health’ of a microbial community. We would like to understand, both biotic and abiotic factors, that are predictive of current and future health of a complex microbial community. Additionally, we aim to decipher the relationship between biodiversity, resistance (ability to maintain functional activity state in face of stress), resilience (ability to recover functional activity state after stressful perturbation), and adaptive capacity (ability to adopt new functional activity states in a new environment) of a microbial community? We pursue these studies by using microbial communities from Oak Ridge Environmental Field Research site and Fluidized Bed Reactors or FBRs. FBRs allow retention of attached populations (thereby retaining non-growth populations and system adaptive capacity, provide non-destructive sampling and can mimic rapid geochemical transitions (e.g. providing for short or prolonged periods of stress). Therefore, FBRs provide excellent system to study health of microbial communities.
