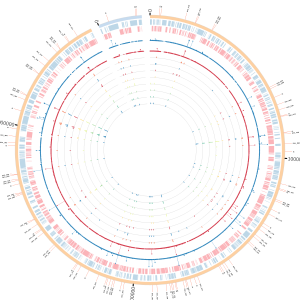
 Previously, a two-organism model community was established under sulfate-deplete conditions that prevented independent growth of Desulfovibrio vulgaris (Dv) by sulfate respiration (SR), and required its syntrophic interaction with Methanococcus maripaludis (Mm) to support growth of both organisms (Hillesland & Stahl, 2010). ‘Syntrophy’ (ST) is an obligate mutualism in which, here, the oxidation of the organic substrate (lactate) by Dv is energetically feasible only if its fermentation products (primarily hydrogen (H2) and formate) are consumed by Mm. Twelve lines of this model community were further subjected to 1000 generations of experimental evolution. Growth characteristics across all 12 lines improved significantly around 300 generations (Hillesland & Stahl, 2010). Whole genome sequencing revealed that many mutations had accumulated in the genomes of both organisms across all 12 lines. Analysis of mutations showed that specialization for ST across nearly all lines came at the expense (tradeoff) of erosion of SR (Hillesland et al, 2014). We are currently investigating if there was any evidence of co-evolution during evolution of syntrophy by using single cell sequencing. Ultimately, we would like to develop a single cell lineage mapping of mutations accumulated to predict emergence and co-evolution of syntrophy. This will also drive development of single cell computational methods and algorithms to build gene regulatory network modeling at the single cell level.
Previously, a two-organism model community was established under sulfate-deplete conditions that prevented independent growth of Desulfovibrio vulgaris (Dv) by sulfate respiration (SR), and required its syntrophic interaction with Methanococcus maripaludis (Mm) to support growth of both organisms (Hillesland & Stahl, 2010). ‘Syntrophy’ (ST) is an obligate mutualism in which, here, the oxidation of the organic substrate (lactate) by Dv is energetically feasible only if its fermentation products (primarily hydrogen (H2) and formate) are consumed by Mm. Twelve lines of this model community were further subjected to 1000 generations of experimental evolution. Growth characteristics across all 12 lines improved significantly around 300 generations (Hillesland & Stahl, 2010). Whole genome sequencing revealed that many mutations had accumulated in the genomes of both organisms across all 12 lines. Analysis of mutations showed that specialization for ST across nearly all lines came at the expense (tradeoff) of erosion of SR (Hillesland et al, 2014). We are currently investigating if there was any evidence of co-evolution during evolution of syntrophy by using single cell sequencing. Ultimately, we would like to develop a single cell lineage mapping of mutations accumulated to predict emergence and co-evolution of syntrophy. This will also drive development of single cell computational methods and algorithms to build gene regulatory network modeling at the single cell level.
