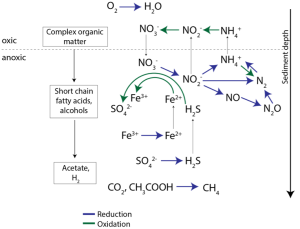
 Nitrate is a key contaminant and electron acceptor and its transformations link together the nitrogen cycle, the carbon cycle, and the fate of various contaminants. By designing environmentally-relevant synthetic communities composed of isolates from the field, we can study nitrate reduction in a laboratory-controlled system. Understanding the nitrogen cycle can shed insight into communities, organisms, processes, genes, and energetics associated with metal reduction.
Nitrate is a key contaminant and electron acceptor and its transformations link together the nitrogen cycle, the carbon cycle, and the fate of various contaminants. By designing environmentally-relevant synthetic communities composed of isolates from the field, we can study nitrate reduction in a laboratory-controlled system. Understanding the nitrogen cycle can shed insight into communities, organisms, processes, genes, and energetics associated with metal reduction.
Our goal is to establish model nitrate-reducing communities in order to elucidate microbial interactions, resilience, and regulatory mechanisms involved in the respiratory process. Previously, we have elucidated how regulatory mutations contribute to resilience of a sulfate-reducing microbial community (Turkarslan et. al., 2017). We plan to apply this developed framework involving systems-level analyses, environment and gene regulatory influence network (EGRIN) models, and single-cell analytics to study nitrate reduction. Individual isolates will first be characterized in pure culture and then communities will be assembled. Intrasporangium calvum C5, an organism with DNRA and denitrification capability, is a current organism of interest and additional isolations targeting relevant organisms are in progress. Fluidized bed reactors (FBRs), more representative of environmental conditions, will eventually be utilized to study nitrate reduction in synthetic communities as well as environmental samples. We will create conditions that select for different physiological states including ammonia production (i.e., DNRA), complete denitrification and incomplete denitrification to N2O. As we have successfully applied for sulfate respiring communities, we will execute transition experiments in FBRs and characterize phenotypic and transcriptomic changes to determine signatures of resilience and adaptive capacity. In addition, FBRs will allow us to perform high-throughput anaerobic enrichments from the field specifically targeting nitrate/sulfate respiring and syntrophically growing communities. These studies will provide a generalized quantitative framework to characterize how anthropogenic activities restructure trophic interactions, to predict consequences of nitrate pollution and its effect on fate of C and to investigate if and how anthropogenic activities have restructured trophic interactions.
