 More than 10 million people fall ill from Mycobacterium tuberculosis infection every year. Putting up a fight against the pathogen is posed with recurrent infections worldwide which is the consequence of resistance and tolerance strains among population. Resistance develop due to prolonged periods of antibiotics treatments which span more than 3 months. Long periods of treatment is required to kill tolerant strains, which as dormant and survive antibiotics which are designed to act on fast growing cells. Thus dormant sub population known as persisters act as a precursor for drug resistance and its essential to study their mechanism of formation to develop better antibiotic treatments. However, several genes and ipso facto diverse mechanisms lead to formation of persisters, making this study a systems biology problem.
More than 10 million people fall ill from Mycobacterium tuberculosis infection every year. Putting up a fight against the pathogen is posed with recurrent infections worldwide which is the consequence of resistance and tolerance strains among population. Resistance develop due to prolonged periods of antibiotics treatments which span more than 3 months. Long periods of treatment is required to kill tolerant strains, which as dormant and survive antibiotics which are designed to act on fast growing cells. Thus dormant sub population known as persisters act as a precursor for drug resistance and its essential to study their mechanism of formation to develop better antibiotic treatments. However, several genes and ipso facto diverse mechanisms lead to formation of persisters, making this study a systems biology problem.
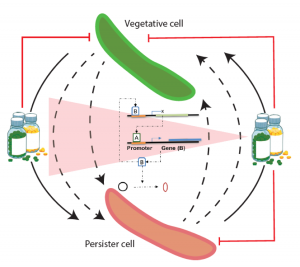
Previously, tolerance of MTB to Bedaquiline was analyzed using system wide model of network architecture (EGRIN), developed by our group. Extending our current expertise in the group, we are developing bioinformatics and experimental framework to reverse engineer the pathways leading to formation of persistence. These network inferences will be used to design antibiotics therapies to lower the tolerance load of the pathogen and understand the emergence of tolerant strains like Beijing genotype.
