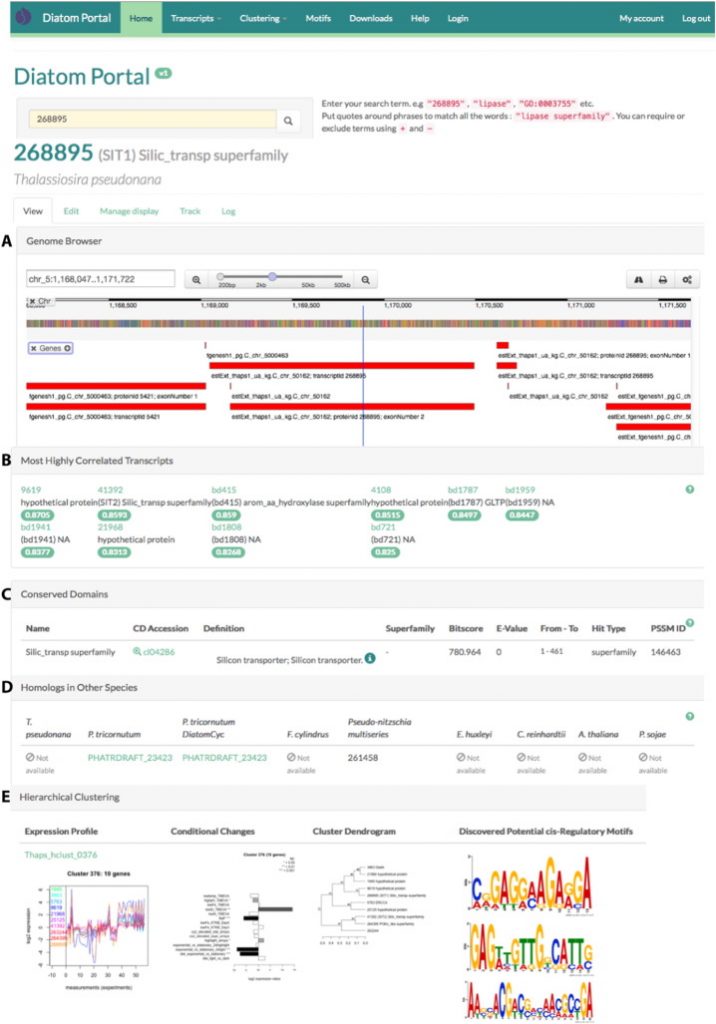
 The Diatom Portal web interface includes gene, transcript and protein level information for fully sequenced and transcriptionally profiled diatoms (currently T. pseudonana and P. tricornutum), a lightweight genome context browser, and the results of hierarchical expression clustering and motif-directed biclustering over many aggregated datasets and conditions. The contents of this portal are organized into gene transcripts, clusters of co-expressed genes, and detected potential DNA cis-regulatory sequence motifs. These can be explored by using a variety of filters, or searched using a variety of search terms including JGI protein identification numbers, annotation keywords, and GO terms. There are two primary record types represented in the portal: Genes and Clusters. Pages for individual genes contain a genome browser, gene and transcript model information, synonymous external identifiers, other most highly correlated transcripts, conserved domains, predicted orthologous genes detected in other diatoms and plant species, previews of co-expression clusters in which the transcript occurs, KEGG mapping and gene ontology (GO) terms.
The Diatom Portal web interface includes gene, transcript and protein level information for fully sequenced and transcriptionally profiled diatoms (currently T. pseudonana and P. tricornutum), a lightweight genome context browser, and the results of hierarchical expression clustering and motif-directed biclustering over many aggregated datasets and conditions. The contents of this portal are organized into gene transcripts, clusters of co-expressed genes, and detected potential DNA cis-regulatory sequence motifs. These can be explored by using a variety of filters, or searched using a variety of search terms including JGI protein identification numbers, annotation keywords, and GO terms. There are two primary record types represented in the portal: Genes and Clusters. Pages for individual genes contain a genome browser, gene and transcript model information, synonymous external identifiers, other most highly correlated transcripts, conserved domains, predicted orthologous genes detected in other diatoms and plant species, previews of co-expression clusters in which the transcript occurs, KEGG mapping and gene ontology (GO) terms.
Pages for individual co-expression clusters contain groups of gene transcripts whose expression was correlated over many experiments, changes in expression over categorical conditions, GO term enrichments, and potential cis-regulatory DNA motifs detected in the upstream promoter regions of the genes occurring in the cluster. The details and position weight matrices (PWMs) for these motifs can also be downloaded in MEME format for comparison against databases of known motifs (e.g. TOMTOM ( Gupta et al., 2007)). The aggregate data set used for expression clustering can also be downloaded for additional or independent analysis. The goal of this Diatom Portal is to facilitate discoveries, hypothesis generation, collaboration and continued data integration within the diatom research community in order to accelerate the advancement of the science and understanding of diatom biology and gene regulation. For example, the incorporation of additional transcriptomic data (including mRNA-sequencing data and the MMETSP (Keeling et al., 2014)) as it becomes available will increase the resolution and conditionality of gene expression clustering, and continued cross-species analysis will expand the set of conserved, related and unique regulatory features and coordination that can be detected and inferred within the diatom clade.
Visit Diatom Portal at: http://networks.systemsbiology.net/diatom-portal
