Futher Investigation: Genomic DNA extraction
Purpose:
To seperate and analyze Halo DNA.
Procedure:
2 mL of a given halo culture was transferred to a microcentrifuge tube and was centrifuged for 2 minutes at maximum speed. The supernatant in the microcentrifuge tube was removed and 500 µL of nuclease free water and 2 µL of 25mg/mL RNase were placed into the microcentrifuge tube. The tube was then incubated in a water bath at 37ºC for 30 minutes.
After removing the tube from the water bath 1000 µL of 100% ethanol was transferred into the microcentrifuge tube and was inverted to mix. The DNA within the tube was picked up with a micropipette tip and placed in 300 µL of 70% ethanol to be washed. The DNA was picked up again and placed in a new empty microcentrifuge tube and spun down. The supernatant was once again removed from the tube and the pellet within the microcentrifuge tube was left to dry for 10 to 15 minutes. 100 µL of TE buffer was then placed in the tube. The tube was vortexed so that no clouds of DNA could be seen.
The nano drop spectrophometer, used to measure the DNA concentration, was cleaned with milli Q water. Nucleic Acid was chosen on the nano drop program and 1 drop of milli Q water was placed on the nano drop. The nano drop was then initialized. Checking to see that the program was set on DNA-50, milli Q water was used to run a blank sample. After cleaning the nano drop again 1 µL of the Halobacterium DNA on the nano drop and the ng/µL was measured and recorded.
In order to continue with the PCR the oliga (gene primers), 25mM dntp, 10x buffer, 5x Q, taq (enzyme) and nuclease-free water were gathered while making sure that both the forward and reverse primers were present for each gene to be amplified. The genes to be amplified using PCR were tbpA, tbpB, tbpC and tbpD. The following table was used to calculate the amount of each substance needed to make a cocktail solution:
0.2 µL 25 mM dntp
2 µL 10x buffer
4 µL 5x Q
1.5 µL Oliga F
1.5 µL Oliga R
0.4 µL taq
+ 10.4 µL H2O
20 µL solution
Since 8 tubes were to be used all the values except the oliga values were multiplied by 9 so as to have extra, thus 1.8 µL of 25 mM dntp, 18 µL of 10x buffer, 36µL of 5x Q, 3.6 µL of taq and 84.6 µL of water were placed into a single microcentrifuge tube and this tube was labeled as “cocktail.” A micropipette was used to transfer 1.5 µL of the tbpA forward primer to a new PCR tube. The same amount of tbpA reverse primer was added to the same tube. After diluting the resuspended DNA so that the concentration is about 50 ng/mL, 1 µL of the resuspended DNA was transferred to the first tube of the PCR tubes. 16 µL of the cocktail was then added to the same tube. This was repeated with the rest of the three primers. The tube was set in the PCR machine and the settings were adjusted accordingly.
While waiting for the PCR to finish the agarose gel to be used for electrophoresis of the genes was made. In order to do this 1 g of agaraose was added to a flaskand 100 mL of TAE buffer was added to the same flask. This solution was then microwaved while checking that the solution didn’t boil over. The microwaving was continued until no bubbles could be seen in the solution. After pouring the solution into a setting that creates the right amount of wells for the number of PCR solutions. The gel was soaked in the electric box with solution for the current after the solution has solidified. When the PCR was finished 2 µL of ethidium bromide was added to each of the PCR tubes. 5 µL of each solution from the PCR tubes was loaded into the wells in the agarose gel while remembering to skip the first and last well. 5 µL of kb DNA ladder was added to the first and last well and the gel was left to run for 24 minutes. Gel doc was used to get a picture.
Results:
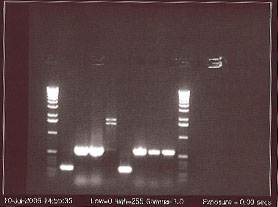
This gel run shows the number of base pairs each gene has. From left to right: DNA ladder, tbpA (215 bp), tbpB (440bp), tbpC (455bp), tbpD (560 bp), tbpA, tbpB, tbpC, tbpD, DNA ladder. tbpA is smaller (has less base pairs) than all of the other genes because it went farther across the gel than tbpB, tbpC, and tbpD did.
[back to top]
|