All our software and resources are open source, and licensed under the GNU Lesser General Public License (LGPL).
cMonkey
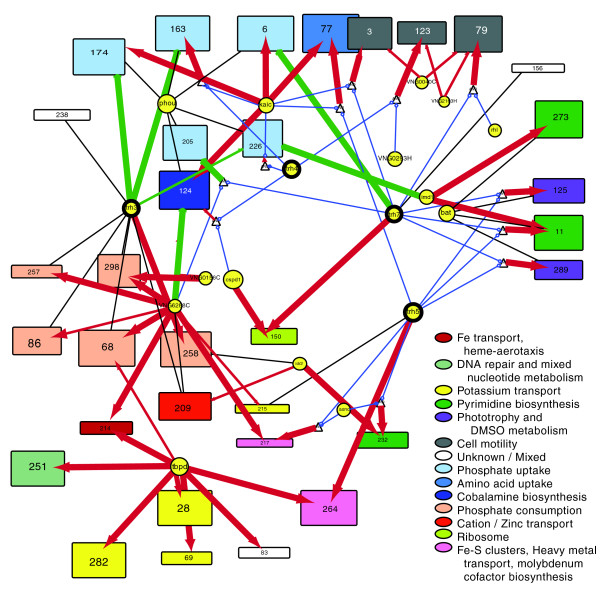
cMonkey detects putative co-regulated gene groupings by integrating the biclustering of gene expression data and various functional associations with the de novo detection of sequence motifs.
cMonkey2
This is the Python implementation of the cMonkey algorithm based on the original R implementation by David J. Reiss, Institute for Systems Biology.
The Inferelator is an algorithm for inferring predictive regulatory networks from gene expression data.
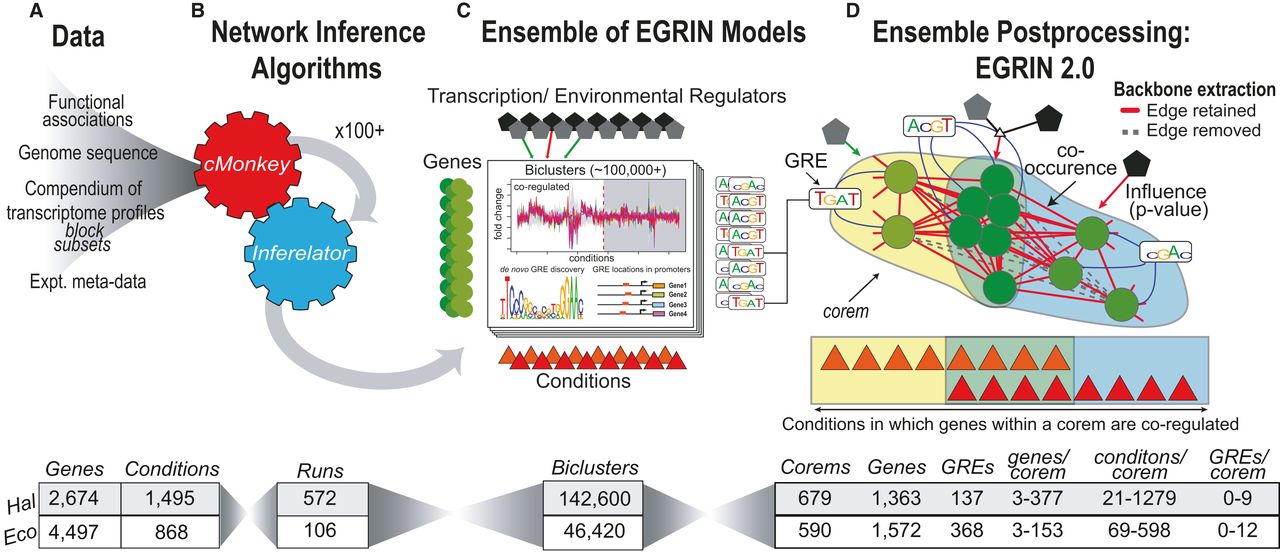
EGRIN2.0
EGRIN 2.0 is a systems-level model that delineates the complex relationship between environment, gene regulation, and phenotype in prokaryotes
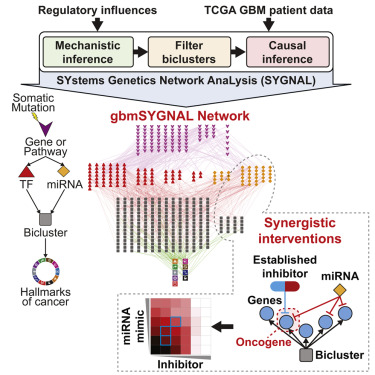
SYGNAL
We developed the SYstems Genetic Network AnaLysis (SYGNAL) pipeline to integrate correlative, causal and mechanistic inference approaches into a unified framework that systematically infers the causal flow of information from mutations to TFs and miRNAs to perturbed gene expression patterns across patients.

gbmSYGNAL
This website was developed to provides easy access to the gbmSYGNAL network by allowing searching of somatically mutated genes, regulators, or genes in biclusters.
MTB Portal
MTB Portal serves as a portal for computational modeling program to generate an integrated, predictive gene regulatory network model of host/pathogen interactions.
The Diatom Portal is a new resource for the diatom community to provide a repository for experimental and computational results, as well as an interface to existing resources.
The Chlamy Portal is a community-wide web-resource to explore the Transcriptional Regulatory Network model for Chlamydomonas reinhardtii.
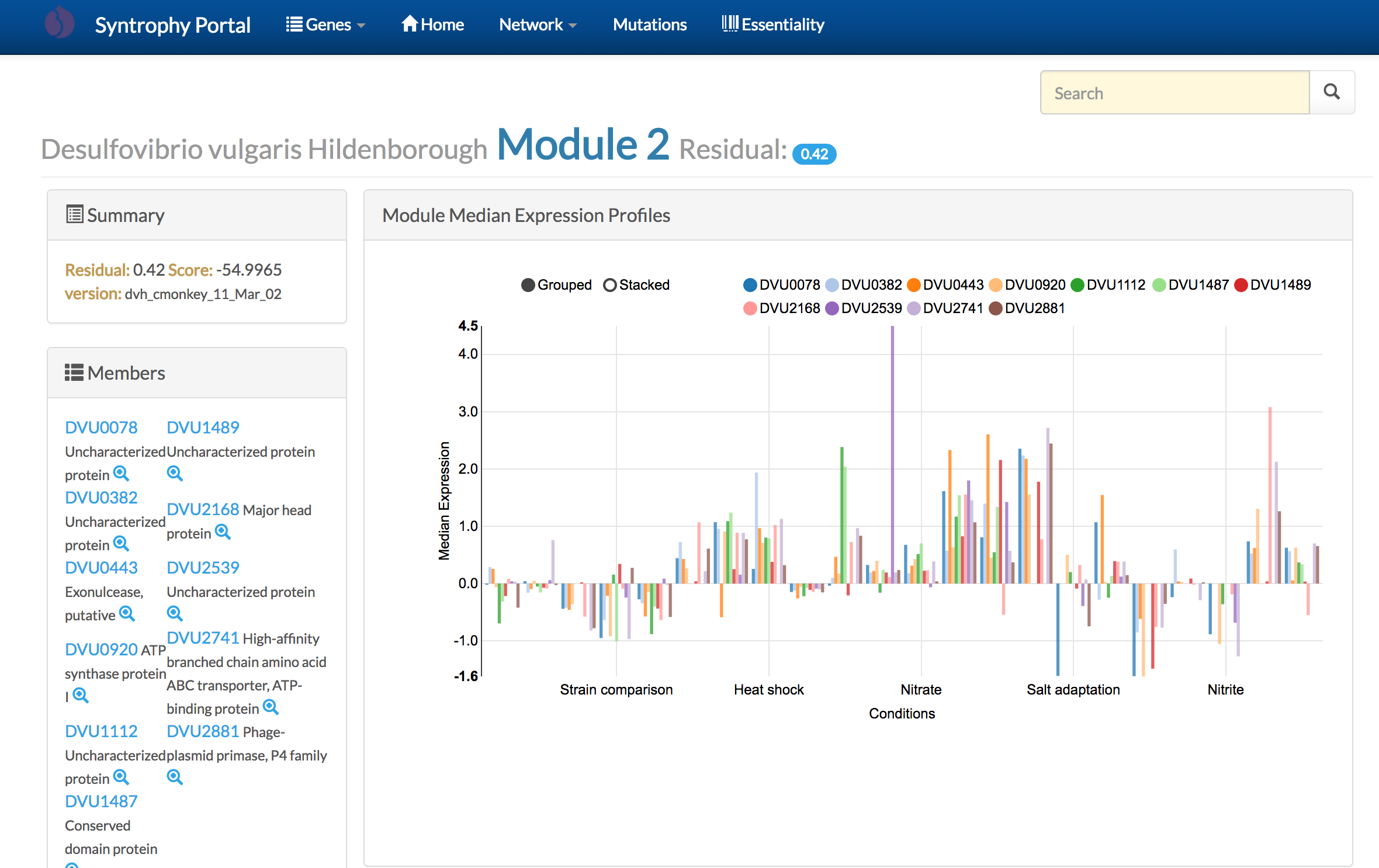
The Syntrophy Portal is a community-wide web-resource to explore the Transcriptional Regulatory Network model for syntrophic model organisms Desulfovibrio vulgaris Hildenborough and Methanococcus maripaludis S2.
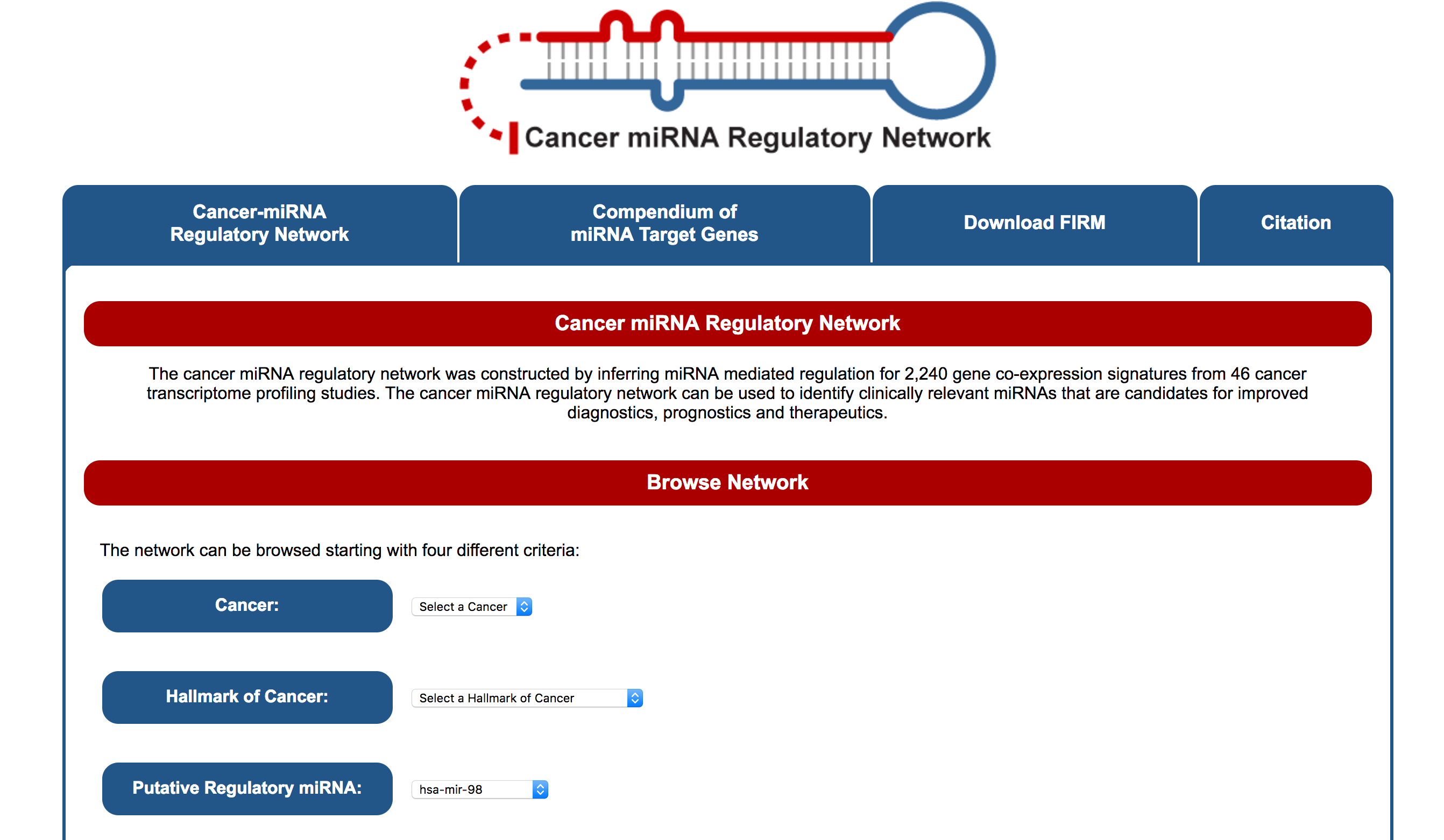
The cancer miRNA regulatory network was constructed by inferring miRNA mediated regulation for 2,240 gene co-expression signatures from 46 cancer transcriptome profiling studies.
Project Feed 1010 aims to educate citizens in sustainable food production and research these systems for optimization.
-
Type
You can search Halobacterium sp. NRC-1 annotations from SBEAMS database.
Halobacterium sp. NRC-1 operon structures Download PDF (74 Mb)