 Glioblastoma multiforme (GBM) is the most common brain tumor and is nearly uniformly fatal. Development of new therapeutics has been slow and difficult (Alexander et al., 2013), in part because GBM is a complex and heterogeneous disease (Brennan et al., 2013). One possible strategy to achieve complete and durable remission is to tailor a combination of drugs that targets multiple vulnerabilities in a patient’s tumor. What is needed to test this strategy is an approach that navigates the large space of possible drug combinations and prioritizes specific drug combinations based on the molecular signatures of a patient’s tumor. We hypothesized that knowledge of the detailed architecture of transcription factor (TF) and microRNA (miRNA) regulatory interactions in the form of a transcriptional regulatory network (TRN) would provide the mechanistic details required to prioritize combinatorial interventions. Both TFs (Cai et al., 1996) and, more recently, miRNAs (Bouchie, 2013) have been used as therapeutic targets. In fact, consistent with the situation ∼20 years ago (Cai et al., 1996), therapies targeting TFs still comprise 14% of the top 50 best-selling FDA-approved drugs in 2014. Additionally, therapies targeting TFs and miRNAs have the potential for a broader effect than those targeting a single gene, as these regulators control many genes associated with diverse oncogenic biological processes.
Glioblastoma multiforme (GBM) is the most common brain tumor and is nearly uniformly fatal. Development of new therapeutics has been slow and difficult (Alexander et al., 2013), in part because GBM is a complex and heterogeneous disease (Brennan et al., 2013). One possible strategy to achieve complete and durable remission is to tailor a combination of drugs that targets multiple vulnerabilities in a patient’s tumor. What is needed to test this strategy is an approach that navigates the large space of possible drug combinations and prioritizes specific drug combinations based on the molecular signatures of a patient’s tumor. We hypothesized that knowledge of the detailed architecture of transcription factor (TF) and microRNA (miRNA) regulatory interactions in the form of a transcriptional regulatory network (TRN) would provide the mechanistic details required to prioritize combinatorial interventions. Both TFs (Cai et al., 1996) and, more recently, miRNAs (Bouchie, 2013) have been used as therapeutic targets. In fact, consistent with the situation ∼20 years ago (Cai et al., 1996), therapies targeting TFs still comprise 14% of the top 50 best-selling FDA-approved drugs in 2014. Additionally, therapies targeting TFs and miRNAs have the potential for a broader effect than those targeting a single gene, as these regulators control many genes associated with diverse oncogenic biological processes.
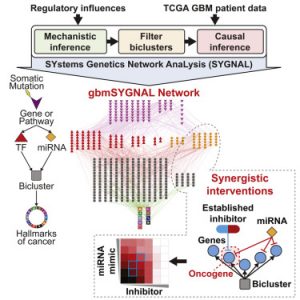
We have developed a platform for integrating somatic mutations and gene expression from patient data into a mechanistically based transcriptional regulatory network. They applied this approach to construct the most comprehensive regulatory network for the deadly brain cancer glioblastoma multiforme (GBM). We used data from a large consortium (TCGA and ENCODE) to develop a highly integrative approach which they demonstrate can be used to discover the underlying topology of regulatory interactions, potential drug targets, and dissect the biological underpinnings of disease traits (e.g. tumor lymphocyte infiltration). For the first time, this network suggests a regulatory mechanism for how increased cellular immune proteins are associated with increased tumor lymphocyte infiltration and decreased patient survival in GBM.
We developed the TF-target gene database (http://tfbsdb.systemsbiology.net) and the SYstems Genetics Network AnaLysis (SYGNAL) pipeline. We have demonstrated that multi-omic data from each patient can be stitched together into a gbmSYGNAL transcriptional regulatory network (http://glioma.systemsbiology.net) to gain clinically/biologically meaningful insights. And that the network structure and integration with orthogonal information (drug targets) can be used to discover intervention points that can lead to synergistic interactions. This demonstrates that ISB’s new SYGNAL pipeline can become a data integration platform that explains the etiology of a disease and provides the knobs which can be turned to maneuver the system back to a healthier state.
