Experimental analysis of TFB family proteins in a halophilic archaeon reveals complex environment-dependent fitness contributions. Gene conversion events among these proteins can generate novel niche adaptation capabilities, a process that may have contributed to archaeal adaptation to extreme environments.
Serdar Turkarslan, David J Reiss, Goodwin Gibbins, Wan Lin Su, Min Pan, J Christopher Bare, Christopher L Plaisier & Nitin S Baliga
Molecular Systems Biology 7:554; doi:10.1038/msb.2011.87; published 22 November 2011
Synopsis | Full Text | PDF | Supp. Inf. | Review Process
Article Highlights
- Evolution of archaeal lineages correlate with duplication events in the TFB family.
- Each TFB is required for adaptation to multiple environments.
- The relative fitness contributions of TFBs change with environmental context.
- Changes in the regulation of duplicated TFBs can generate new adaptation capabilities.
Synopsis
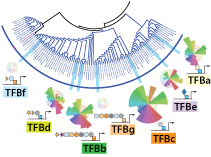
The evolutionary success of an organism depends on its ability to continually adapt to changes in the patterns of constant, periodic, and transient challenges within its environment. This process of ‘niche adaptation’ requires reprogramming of the organism’s environmental response networks by reorganizing interactions among diverse parts including environmental sensors, signal transducers, and transcriptional and post-transcriptional regulators. Gene duplications have been discovered to be one of the principal strategies in this process, especially for reprogramming of gene regulatory networks (GRNs). Whereas eukaryotes require dozens of factors for recruitment of RNA polymerase, archaea require just two general transcription factors (GTFs) that are orthologous to eukaryotic TFIIB (TFB in archaea) and TATA-binding protein (TBP) (Bell et al, 1998). Both of these GTFs have expanded extensively in nearly 50% of all archaea whose genomes have been fully sequenced. The phylogenetic analysis presented in this study reveal lineage-specific expansions of TFBs, suggesting that they might encode functionally specialized gene regulatory programs for the unique environments to which these organisms have adapted. This hypothesis is particularly appealing when we consider that the greatest expansion is observed within the group of halophilic archaea whose habitats are associated with routine and dynamic changes in a number of environmental factors including light, temperature, oxygen, salinity, and ionic composition (Rodriguez-Valera, 1993; Litchfield, 1998).
We have previously demonstrated that variations in the expanded set of TFBs (a through e) in Halobacterium salinarum NRC-1 manifests at the level of physical interactions within and across the two families, their DNA-binding specificity, their differential regulation in varying environments, and, ultimately, on the large-scale segregation of transcription of all genes into overlapping yet distinct sets of functionally related groups (Facciotti et al, 2007). We have extended findings from this earlier study with a systematic survey of the fitness consequences of perturbing the TFB network of H. salinarum NRC-1 across 17 environments. Notably, each TFB conferred fitness in two or more environmental conditions tested, and the relative fitness contributions (see Table I) of the five TFBs varied significantly by environment. From an evolutionary perspective, the relationships among these fitness landscapes reveal that two classes of TFBs (c/g- and f-type) appear to have played an important role in the evolution of halophilic archaea by overseeing regulation of core physiological capabilities in these organisms. TFBs of the other clades (b/d and a/e) seem to have emerged much more recently through gene duplications or horizontal gene transfers (HGTs) and are being utilized for adaptation to specialized environmental conditions.
We also investigated higher-order functional interactions and relationships among the duplicated TFBs by performing competition experiments and by mapping genetic interactions in different environments. This demonstrated that depending on environmental context, the TFBs have strikingly different functional hierarchies and genetic interactions with one another. This is remarkable as it makes each TFB essential albeit at different times in a dynamically changing environment.
In order to understand the process by which such gene family expansions shape architecture and functioning of a GRN, we performed integrated analysis of phylogeny, physical interactions, regulation, and fitness landscapes of the seven TFBs in H. salinarum NRC-1. This revealed that evolution of both their protein-coding sequence and their promoter has been instrumental in the encoding of environment-specific regulatory programs. Importantly, the convergent and divergent evolution of regulation and binding properties of TFBs suggested that, aside from HGT and random mutations, a third plausible (and perhaps most interesting) mechanism for acquiring a novel TFB variant is through gene conversion. To test this hypothesis, we synthesized a novel TFBx by transferring TFBa/e clade-specific residues to a TFBd backbone, transformed this variant under the control of either the TFBd or the TFBe promoter (PtfbD or PtfbE) into three different host genetic backgrounds (Δura3 (parent), ΔtfbD, and ΔtfbE), and analyzed fitness and gene expression patterns during growth at 25 and 37°C. This showed that gene conversion events spanning the coding sequence and the promoter, environmental context, and genetic background of the host are all extremely influential in the functional integration of a TFB into the GRN. Importantly, this analysis suggested that altering the regulation of an existing set of expanded TFBs might be an efficient mechanism to reprogram the GRN to rapidly generate novel niche adaptation capability. We have confirmed this experimentally by increasing fitness merely by moving tfbE to PtfbD control, and by generating a completely novel phenotype (biofilm-like appearance) by overexpression of tfbE.

