Turn on the radio, and a marvelous thing happens. Intricately wired circuits come to life and, as if by magic, the speakers send out music. Of course, you protest, it’s not magic, as a quick look inside can show.
Live Wires
A New Computer Model Sheds Lights On The Complex Networks In A Cell
 |
Switch the wires around, measure the current, and add and remove transistors, batteries, and circuit boards (don’t forget to take notes) and sooner or later, with some deduction and analysis, you can figure out what each of the parts do.
But try the same thing with a living organism and usually the approach fails miserably. Yet this is essentially what Nitin Baliga at Seattle’s Institute for Systems Biology and Rich Bonneau at New York University (NYU) did earlier this year, when they developed a remarkably accurate model of the salt-loving bacteria, Halobacterium salinarum. The model, which appeared in the journal Cell, uses the tiny creature’s genome and just a few measurements, but it can accurately predict the changes in gene expression when the microbe is exposed to various toxic metals, differing levels of oxygen or light, and even different genetic mutations.
Predictions like these will be critical to scientists who design bacteria to make potent anti-malarial drugs or to eat toxic waste. It might also help in the design of salt-resistant crops.
It turns out that even simple predictions, such as what the microbe will do when exposed to more oxygen but less light, are very hard to get right. Although over 500 genomes have been sequenced, they provide frustratingly incomplete information. The problem is that even though the genes direct the synthesis of thousands of RNA and proteins, those molecules don’t just sit there as the environment changes. Proteins and RNA turn each other on and off through feedback loops so intricate they tend to confound biologists, especially those who try to take them apart like radios.
"Characterizing the function of individual proteins and genes tells you a lot about biochemistry and physiology,” says Baliga. "But it is not going to tell you about how the cell comes together to operate.”
Consider this simple experiment. Shine light on a microbe like H. salinarum, and it will respond by making hundreds of light-harvesting proteins to turn the light into energy. But it will also make other proteins called transcription factors, which actively turn on or shut down a variety of genes in the cell.
"The cell has only a limited amount of energy and resources, and it can’t do everything,” says Baliga. "It needs to balance the operation of different processes so it can optimally use its resources.” The way the cell responds to changes is controlled by informational circuits that have proven frustratingly elusive.
For this reason, mapping a cell as unfamiliar as H. salinarum shows the power of the team’s approach. Little was known about this unusual microbe beyond its noteworthy resistance to radiation, and its strange desire for levels of salt that would kill a normal cell. It is commonly found living on sun-blasted landscapes such as the Dead Sea, and can survive totally encased in salt.
 |
To get insight into its exotic biology, the research team started with its genome. Many of the genes were relatively easy to assign function to, since those genes also appeared in other organisms such as E.coli, which have been studied for years.
"For most of the core functions, like how they utilize sugars, and how they make energy, and how they make amino acids,” says Baliga, "the sequences are very similar.”
In other cases, the genes were unfamiliar, and figuring out what they did was much harder. On the assumption that the unknown genes might partially resemble previously discovered genes, the team used sophisticated software originally developed by Bonneau and colleagues in David Baker’s lab at the University of Washington to match the new genes to the old.
The next task was trying to build a "circuit diagram” of the cell. While this was the hardest part of the project, it also held the most promise. Having such a living map of the cell would teach the team how the cell balanced certain processes. In the future, such circuit diagrams could help scientists engineer bacteria to make cheap pharmaceuticals, consume toxic waste, or produce energy.
Mapping the cell meant performing a few hundred classic molecular genetics experiments. The team exposed the cells to 10 different environmental factors, such as light, oxygen, and various toxic metals, and also changed 32 different genes, all in different combinations. Then they took notes.
After 256 experiments, the biologists had an enormous pile of data, much of it interconnected and somewhat redundant. Many of the genes, it turned out, did not operate individually, but instead as members of larger gene groups. Each group was different from the rest, but still connected by networks of transcription factors that controlled the eventual production of proteins. The relationships were dense, interdependent, and complex.
This is where the systems biology approach, which often connects scientists from different fields, paid off. Scientists in Baliga’s group at ISB worked with Rich Bonneau’s team at New York University to create machine learning algorithms (that is, computers that teach themselves) that took data from the initial experiments and created a model of the organism.
The payoff? When the combined team asked their computer model to predict the outcomes of over 147 real-life environmental changes, none of which had yet been previously studied, the model made accurate predictions for up to 80 percent of the microbe’s genes.
According to a press release by Bonneau, understanding how such biological systems function can focus research on engineering the biosynthesis of pharmaceuticals and biofuels.
 |
"The flip side of systems biology is synthetic biology, in which people build circuits. To me they are complementary fields,” adds Baliga.
For instance, a modified version of the "salt-resistance circuit” in H. salinarum might one day help to make corn plants more salt-resistant. The main obstacle is correctly wiring the circuit into the hundreds of more complicated circuits in the different types of corn cells. A predictive model of corn cells could therefore hasten progress toward a salt-resistant plant.
While a project modifying corn has yet to take off, such technology may not be as outlandish as it appears. A team of synthetic biologists at Berkeley led by Jay Keasling, for instance, recently created a modified circuit of 20 genes in E. coli that increased the production of a building block to artemisinin, the most potent anti-malarial drug known, by over 10,000 fold.
Still, biology is full of surprises. Recently, biologists in Barcelona and Germany started switching around the wires in E. coli , measuring the proteins, and recombining parts of genes with other parts, just to see what would happen. Their work appeared in the journal Nature in April. To their surprise, the bacteria tolerated many of the changes without much complaint.
Try the same thing with a radio, and usually the approach fails miserably.
Jake Ashcraft is a graduate student in chemistry at the University of Washington
Images:
Top
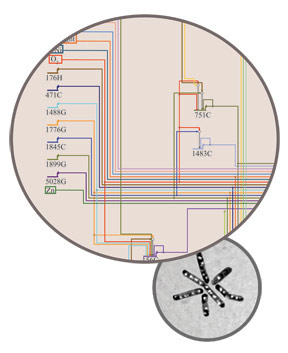
Large circle: A diagram of the genetic networks uncovered by the model. Small circle: Halobacterium salinarum.
Middle: Nitin Baliga
Bottom: Nitin Baliga (left) and some of the team hard at work.
Photos: Institute for Systems Biology

