A Note From the Interns
Thank you to our mentor, Claudia Ludwig, for her support and relentless encouragement. Thank you to the ST-Analytics team for inviting us to their meetings so we could learn more about the world of science. Our teachers here at ISB, Sara Calder, Miranda Johnson, James Park and Jasmine Juette, appreciate all the times you've come to our aid when we were lost. Finally, thank you to the free banana cart from Amazon and the fancy coffee machine on the 4th floor, constantly supplying us with hot chocolate on a breezy summer day.
This summer, Layla Ismail and Suwayda Said worked with various cancer datasets procured by genomics, clinical, and proteomics data. Both interns developed a clear understanding of systems biology by working with diverse data.
One data source came from Glioblastoma cells. Glioblastoma (GBM) is a highly complex and lethal cancer with a 10% chance of survival after five years. Due to this, scientists have devoted countless efforts to analyzing GBM and what makes up its tumor microenvironment. By analyzing the cell heterogeneity using scRNA-seq data (a sequencing method that allows us to look at gene expression), we can examine the efficacy of available GBM treatments.
This summer, Layla worked on completing, optimizing, and publishing two learning modules using research data from Glioblastoma cells. One module was an extension of an activity made by James Park. It focused on characterizing Glioblastoma cell heterogeneity in response to drug treatment. Through this, both interns learned to analyze the efficacy of two drug treatments compared to the control subpopulation. Layla gained inspiration from this and opted to create an advanced module focusing on stem cell marker expression in Glioblastoma tumor samples in R. She analyzed the function of stem cell markers and the cell proliferation pathway they are associated with. Both learning modules can be accessed here:
-
Characterizing GBM Cell Heterogeneity in Response to Drug Treatment
-
Analyzing the Presence of Glioblastoma Stem Cell Markers in the Tumor Microenvironment
She also worked with Jasmine Juette on using data processing to analyze Tumor Mutation Burden. Tumor Mutation Burden is the number of somatic mutations per megabase of the sequenced tumor genome. Tumor cells with high mutation burden effectively fight off immunotherapy, making it extremely difficult to block their cell proliferation pathway. TMBs are used as a biomarker to see how patients are reacting to the immunotherapy given. She worked with a clinical dataset filled with information from 1600+ patients. The outcome of this analysis was recognizing the effect of TMB on the survival rate of patients in this dataset and building a machine-learning model that reflected the original data.
Layla and Suwayda also worked on a National Cancer Institute (a subdivision of the NIH) project with the ST-Analytics team at ISB. This project focuses on analyzing sequential immunotherapies as cancer treatment options as compared to monotherapies or non-sequential combination therapies. They collaborated with scientists from Baliga Lab, Shmulevich Lab, Heath Lab, Wei Lab, and researchers from Yale and UCLA. The goal was to understand this multi-institutional project to help others learn about innovative immuno-oncology systems biology research.
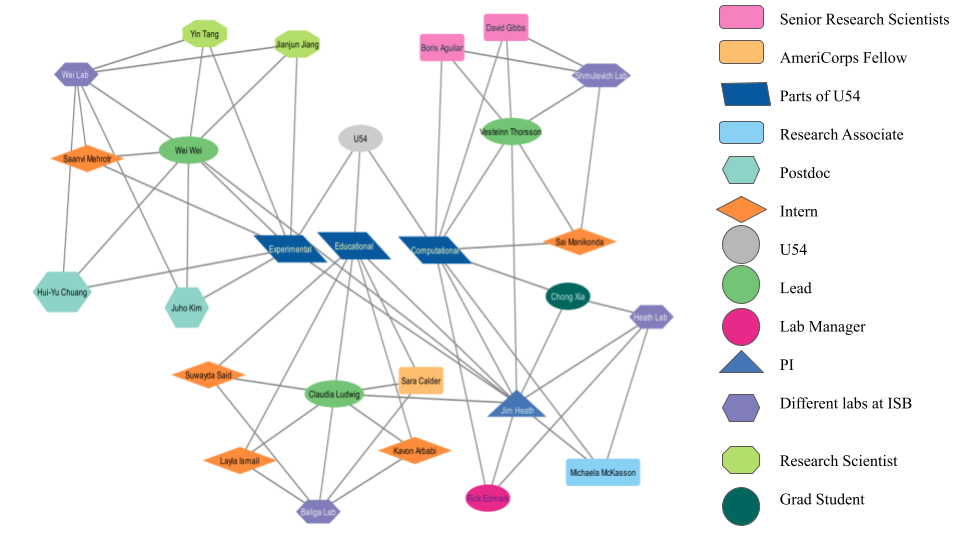
One subgoal was to learn about the 20+ people who have come together to work on this project over the next five years. The intern pair interviewed nine people who are part of the ISB team and created this map, using the tool Cytoscape, which showcases the connections between postdocs, interns, leads, PIs, etc, at ISB. They thought showcasing the wide range of experience, skills, and education across the ST-Analytics team at ISB and everyone working on the U54 NIH project would be essential. It's important to note that this only showcases 1 of the three institutions working on U54, so an actual map of everyone working on the project is even more complex.
During the interviews, Layla and Suwayda gained insight into potential careers. They received fantastic advice especially relevant to young adults interested in science. Thank you to all who gave their precious time for the benefit of students. Here is a word cloud demonstrating specific pieces of wisdom.


