A Note From the Interns
Amelie and Selin would like to acknowledge their mentors Drs. Jake Valenzuela & Chris Deutsch and undergrad intern Brianna Terry for their invaluable guidance and support throughout each step of the research process. In addition, thanks to Karl Gassier for setting up the qPCR thermocycler and making the reaction possible.
Thank you to Claudia Ludwig, Sara Calder, and Miranda Johnson for all their support during the application process and remainder of the internship. Thank you to Dr. Baliga, the Baliga Lab, and all of ISB for welcoming them and providing the opportunity to have an authentic research experience this summer.
Finally, a big thanks goes to their fellow interns for making this the fun and unforgettable experience it was!
Project Overview
CONTEXT
Diatoms are unicellular phytoplankton (microalgae) that play a pivotal role in both the oceanic and atmospheric carbon cycles. Their distinguishing feature is a cell wall crafted from hydrated silica called a frustule which manifests in an array of beautiful patterns. Generating 20-40% of the total oxygen produced on earth and accounting for ~40% of marine primary productivity, diatoms are crucial for carbon sequestration and ocean ecosystems (Armbrust, 2009). Therefore, the activities of diatoms have a dramatic cascading effect on the rest of the biosphere and are important to study as climate change brings many transformations like ocean acidification.
Ocean acidification occurs when excess amounts of CO₂ are trapped in the atmosphere and absorbed by the ocean. When CO₂ combines with water, it forms carbonic acid, which subsequently breaks down into bicarbonate and hydrogen ions. The hydrogen ions lower the water’s pH and disrupt ocean ecosystems (Barker & Ridgwell, 2012). These changes also affect various industries and economies which depend on oceanic productivity (Liou, 2022). By the end of the 21st century, ocean pH levels are expected to decrease by 0.3-0.4 units, which translates into a 100-150% increase in acidity (Liou, 2022).
In these future acidified ocean conditions, previous studies have shown a unique increase in the resilience of diatoms. This increased resilience is due to the diatoms’ ability to more efficiently switch between different cell states in response to stress, which can be studied by looking at changes in the RNA transcriptome under different environments (Valenzuela et al., 2018).
THAPS at 10,000X Magnification

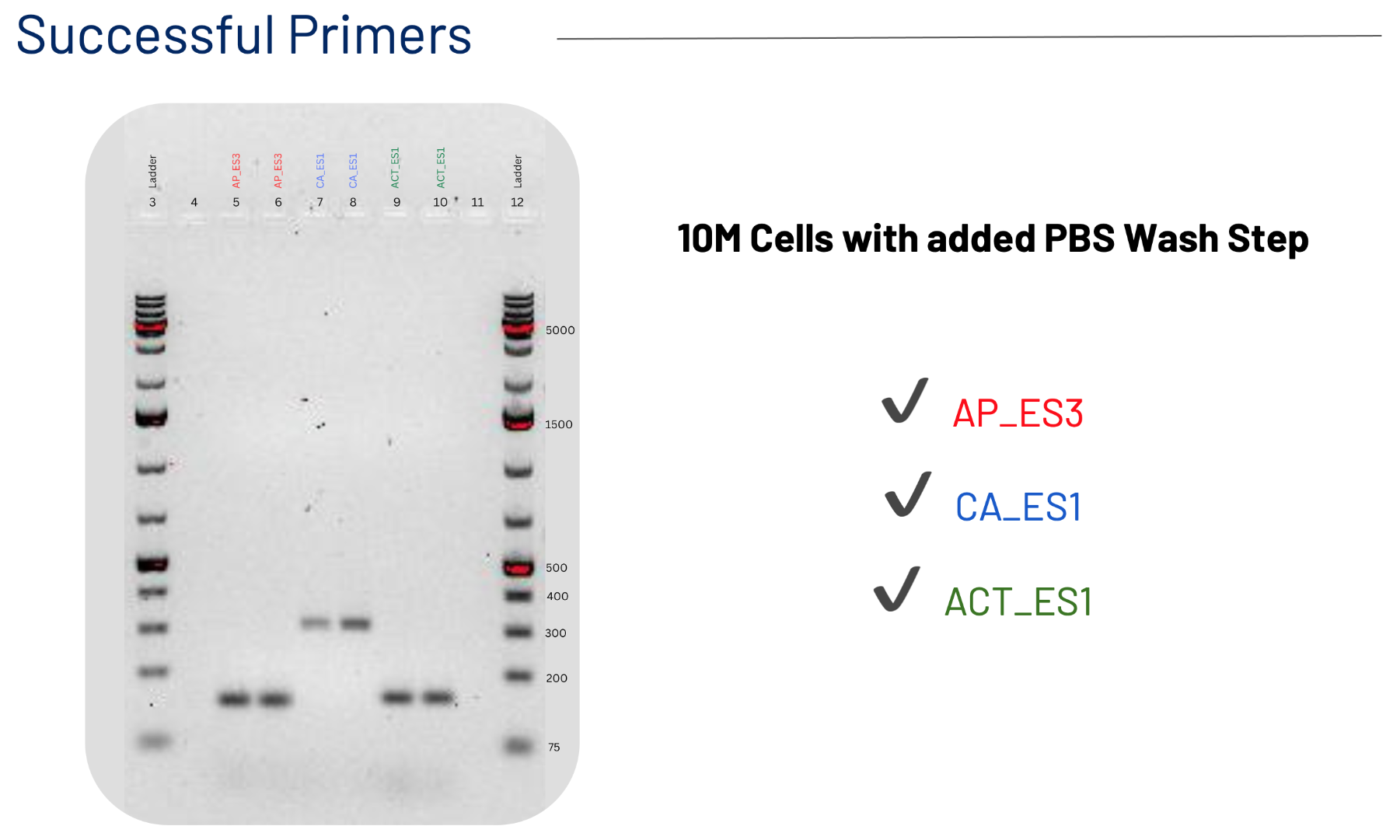
Preparing Primers

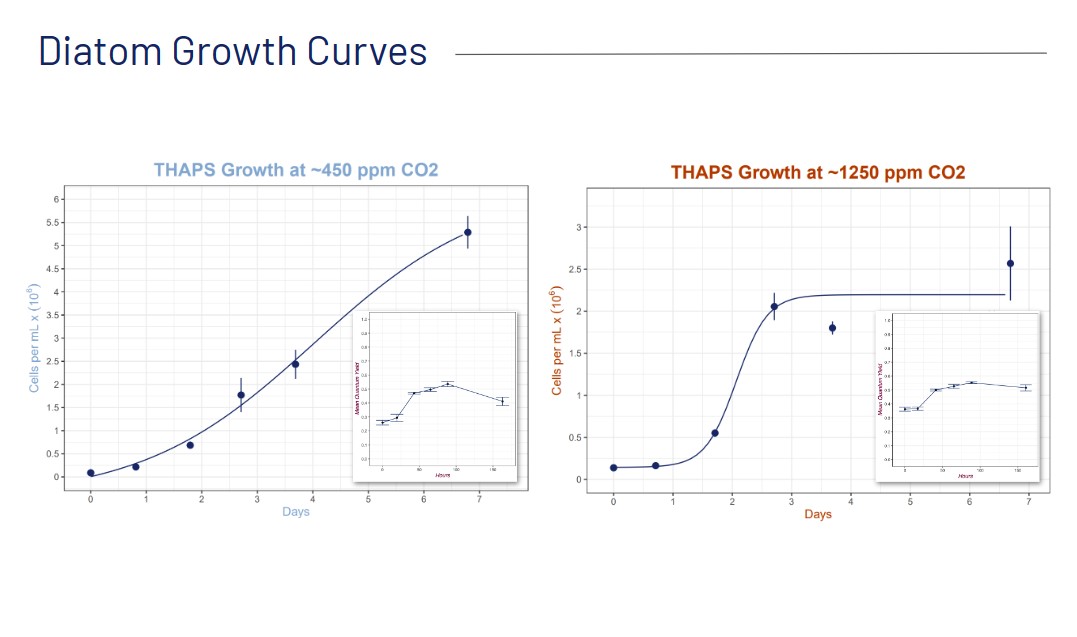
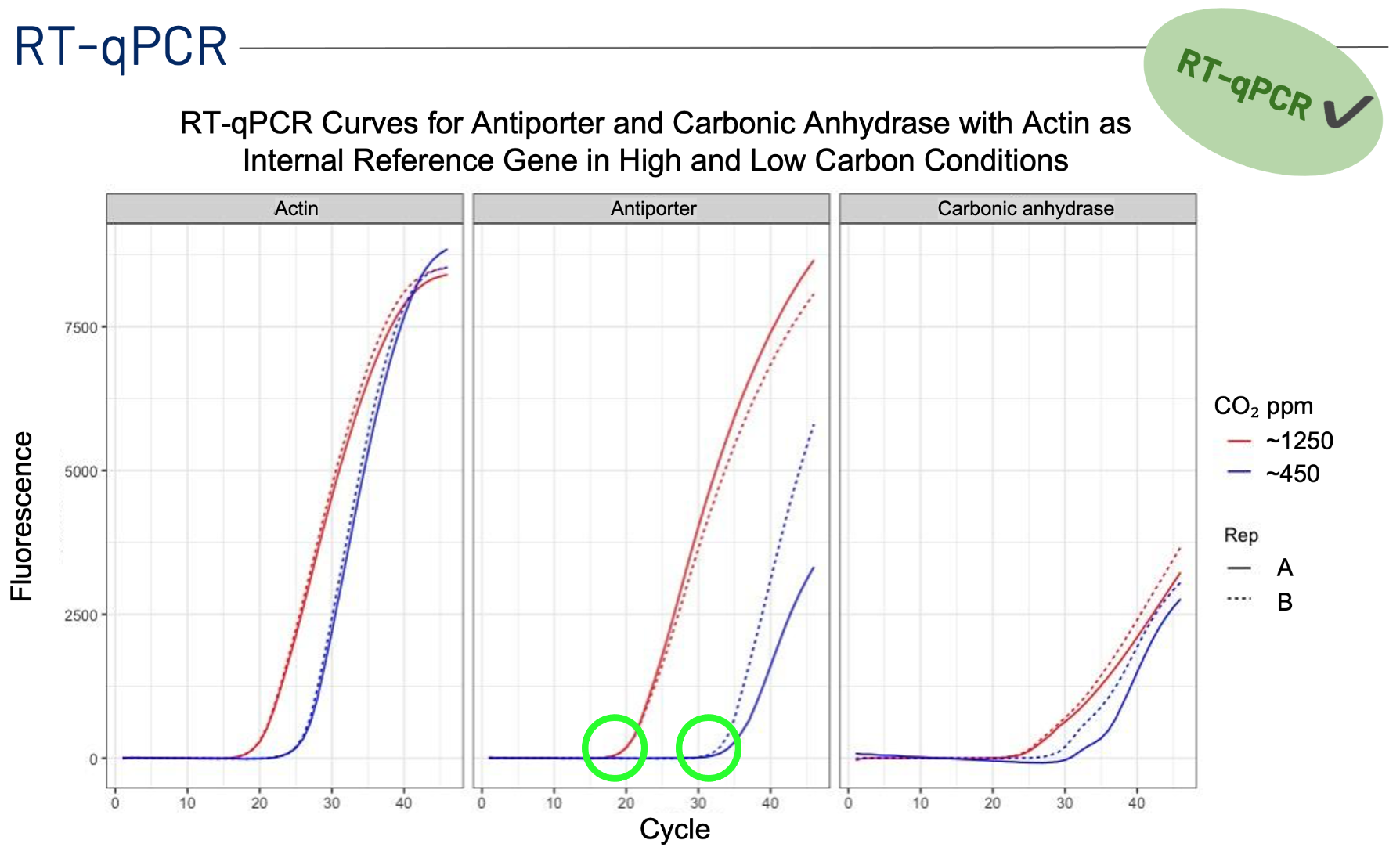
Previous studies identified the genes Na+/H+ antiporter 262258 and carbonic anhydrase 233 as two potential biomarkers to predict a diatom’s transition from low to high carbon states, since both genes are shown to be differentially expressed in high and low carbon conditions. The antiporter is expressed more, or upregulated, in high carbon conditions to maintain the pH of the cell's chloroplasts and expressed less (downregulated) in low carbon. On the contrary, carbonic anhydrase is downregulated in high carbon conditions and upregulated in low carbon, where it plays a crucial role in actively transporting carbon for photosynthesis across the cell wall (Valenzuela et al., 2021). The expression levels of these two genes point to the carbon state a diatom is in, and therefore can serve as “biomarkers” for contemporary (~400 ppm CO2) and future acidified (~1200 ppm CO2) ocean carbon conditions.
To better understand and predict the impacts of ocean acidification on diatoms in future acidified ocean conditions, Amelie and Selin were given the opportunity to work on a project under Drs. Jake Valenzuela and Chris Deustch in the Baliga lab.
AIM
The aim of Amelie and Selin’s project was to pilot a reverse-transcription quantitative PCR or RT-qPCR assay for model diatom T. pseudonana (THAPS) carbon-state biomarkers antiporter 262258 and carbonic anhydrase 233 with actin 260856 as an internal reference gene.
Previously, metatranscriptomics were used to understand the transcriptomic state changes in diatoms and to identify differentially expressed genes. However, metatranscriptomics considers the data for all the genes of all the organisms in a sample. Because of this, highly expressed genes dominate and conceal lowly expressed genes that play important roles in diatom physiology. An RT-qPCR can cut through all this noise and target these low expression genes, like the antiporter and carbonic anhydrase, much more efficiently and inexpensively. With this assay, the hope is to later develop models that can efficiently predict the carbon state of diatoms by quantifying these biomarkers. In the ocean, these biomarkers can serve as an early warning sign that oceanic ecosystems are changing in response to rising CO₂ levels.
Amelie and Selin’s project centered around designing RT-qPCR primers, growing and sampling diatoms in different carbon conditions (contemporary ~450 ppm CO2 and future acidified ~1250 ppm CO2), using the coding language R to visualize data in plots, and developing and testing protocols for RNA and DNA extraction, reverse-transcription, PCR, gel electrophoresis, and RT-qPCR.
RNA TRIzol Extractions