Note from the Interns
Thank you so much to our mentor, Dr. Eric Deutsch, for the immense guidance and support throughout our entire internship. We are also incredibly grateful to Claudia Ludwig and the SEE Team for all of their support this summer. Thank you to the Moritz Lab, the fourth floor, to all of ISB, and to the Amazon community banana stand for such a welcoming environment. Finally, thank you to our fellow interns for all the laughs we shared.
Sincerely,
Clarissa & Marie
The Problem
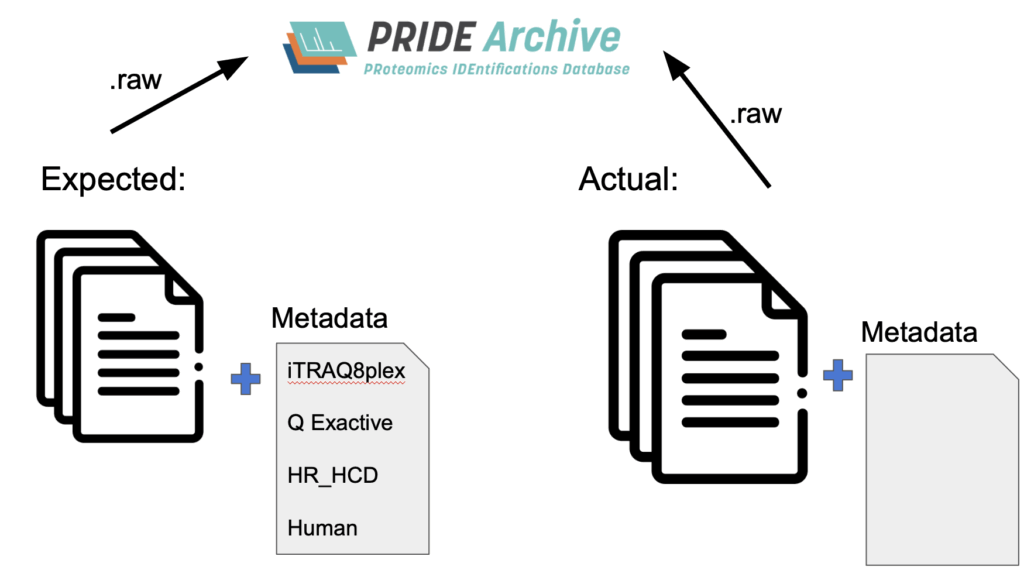
Deposition of mass spectrometry (MS) based proteomics datasets into public data repositories has become widespread due to the efforts of the ProteomeXchange Consortium in cooperation with many journals. This has led to the deposition of over 67,000 datasets, of which over 45,000 are already publicly released. Such huge data availability promotes the reuse of publicly available datasets for a wide range of uses, including training of AI models and building compendia of the detected proteomes of various species (REFs). Such widespread reuse amplifies the value of the vast amount of MS-based proteomics data generated by research projects everywhere.
However, the metadata associated with deposited datasets is often very minimal and thus the information needed to successfully reprocess the data is not easily available. Most of the information can be extracted by reading the original articles that accompany the data deposition, but closed access articles and occasional errors in the published methods contribute to uncertainty in how to specify optimal reprocessing parameters. Yet, the raw data files themselves can reveal many optimal reprocessing parameters via clever programmatic evaluation in most cases.
Our Solution
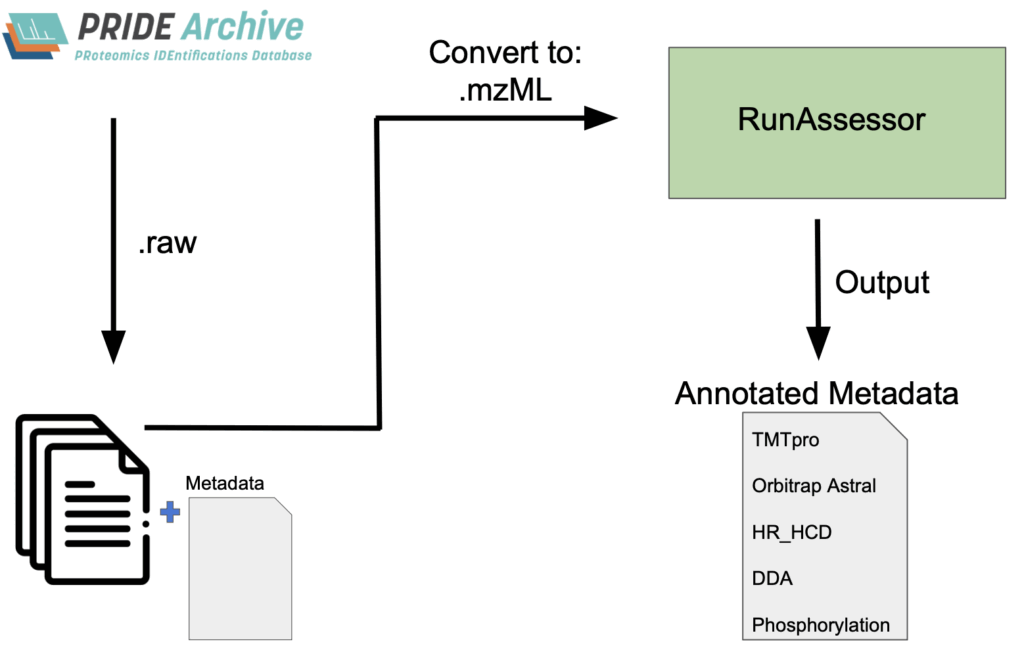
RunAssessor is a command-line software implemented in Python that systematically extracts and summarizes metadata directly from mass spectrometry files. In addition to identifying instrument and acquisition parameters, RunAssessor detects key sample modifications introduced prior to data acquisition, including chemical tagging and phosphorylation enrichment.
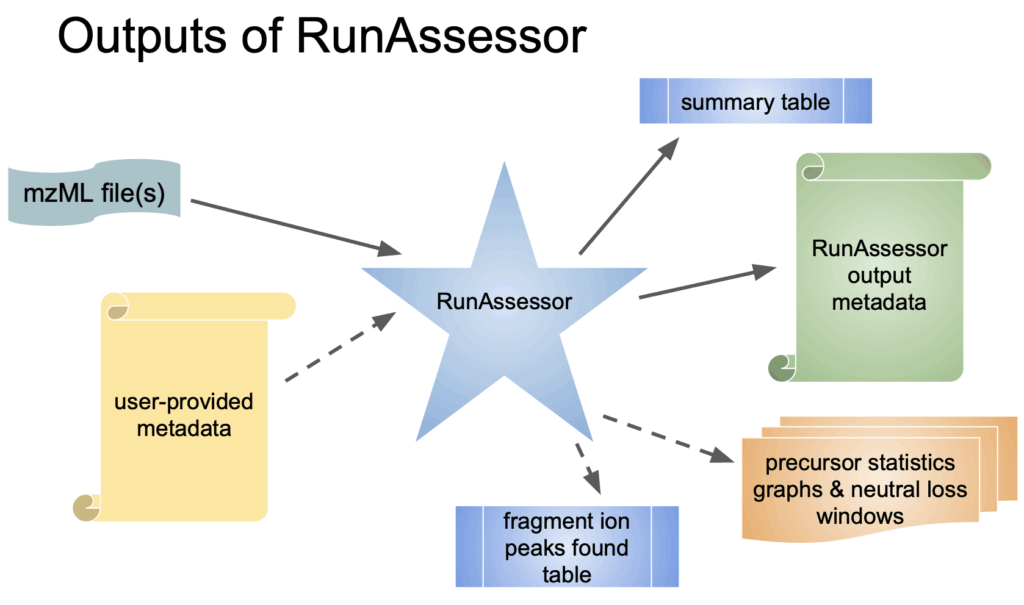
RunAssessor generates two complementary outputs. First, it produces a JSON file that records all fitted low-end and neutral-loss peaks identified in the spectra, along with precursor statistics and direct annotations of any chemical tagging detected. Second, it provides a TSV summary table, which distills the JSON data into a high-level overview. This summary highlights recommended fragmentation methods and precursor tolerance values appropriate for the instrument used to acquire the data.
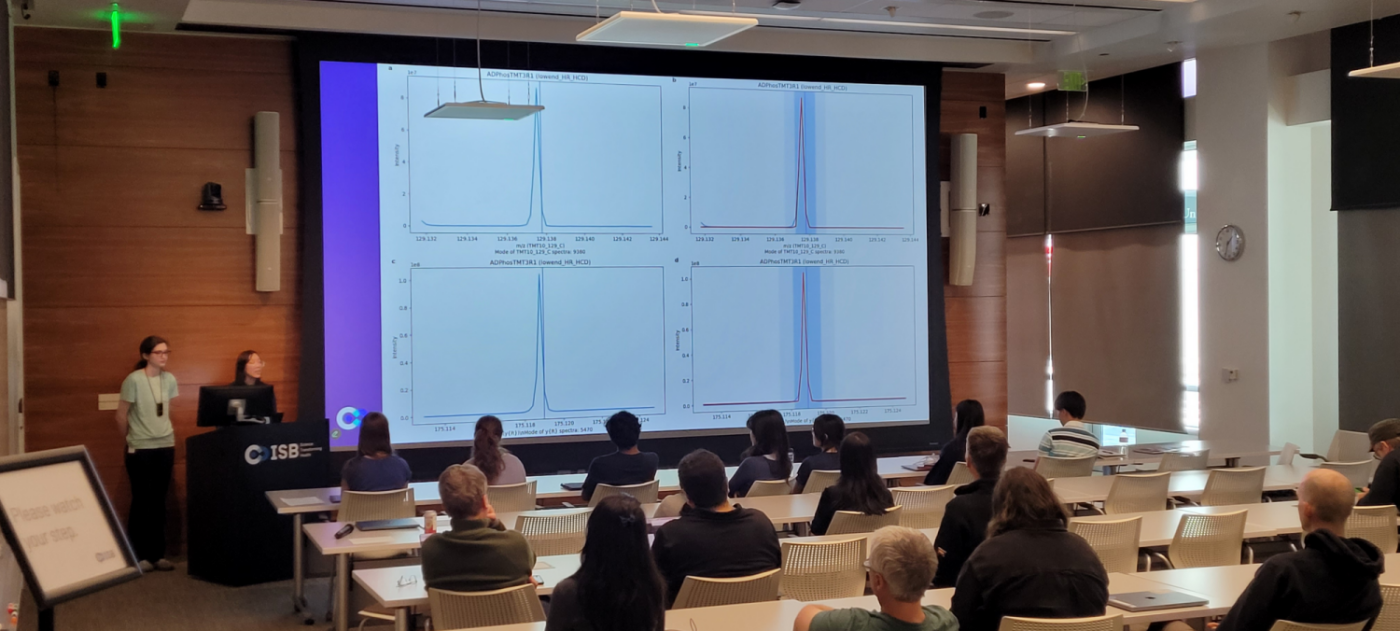
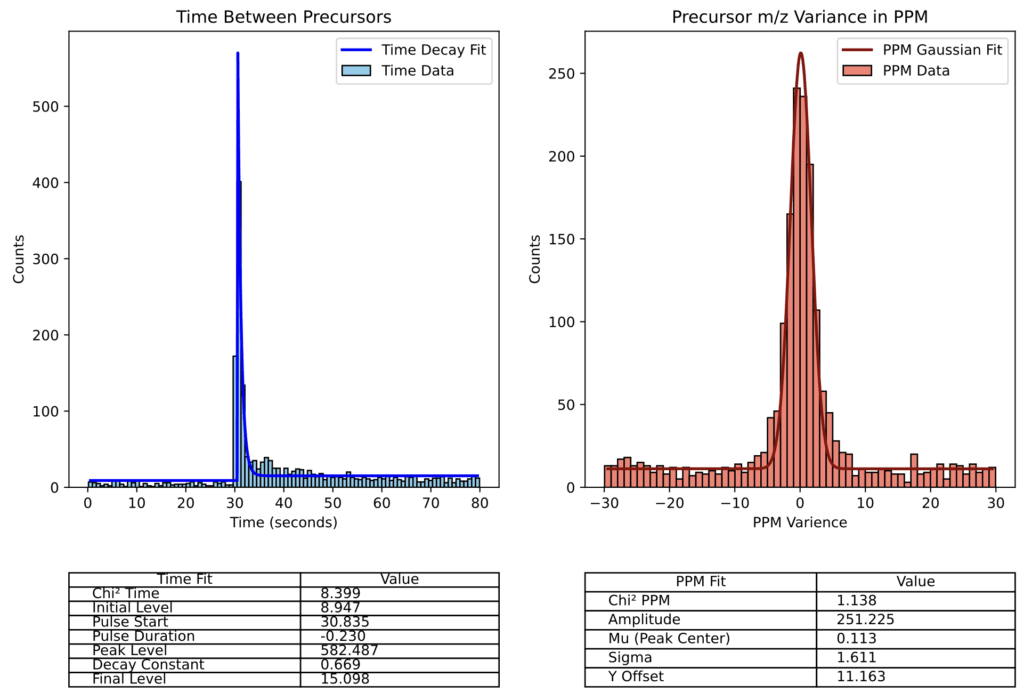
Graphical Outputs
You can also choose to have RunAssessor print out graphs showing the curve fits used for finding the dynamic exclusion time and precursor tolerance.
As well as graphs that show the neutral loss peak fits including 2+ water loss and 2+ phosphoric acid.