Effects of Storage Conditions on Lipid Nanoparticles (LNPs)
Context
Extracellular vesicles (EVs) are lipid membrane-bound nanoparticles secreted by all cells that transport material such as DNA from source to recipient cells. EVs are valuable to study because they can help identify biomarkers. Additionally, they are a less invasive and more accurate way of diagnosing different diseases and disorders and transporting medicine.
Lipid nanoparticles (LNPs) are artificial vesicles that can carry molecules like DNA in their core. This storage experiment aimed to analyze the DNA cargo’s stability under different storage conditions for future research and therapeutic purposes.
Experimental Plan
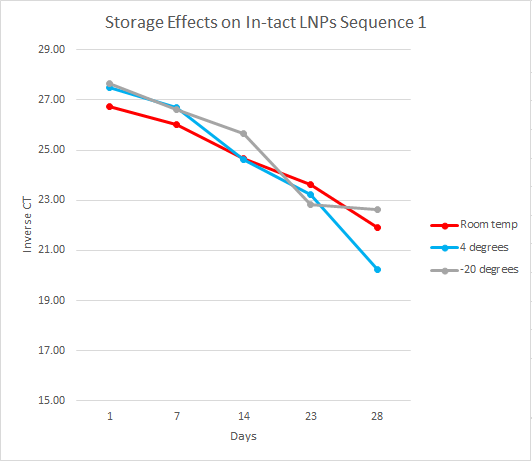
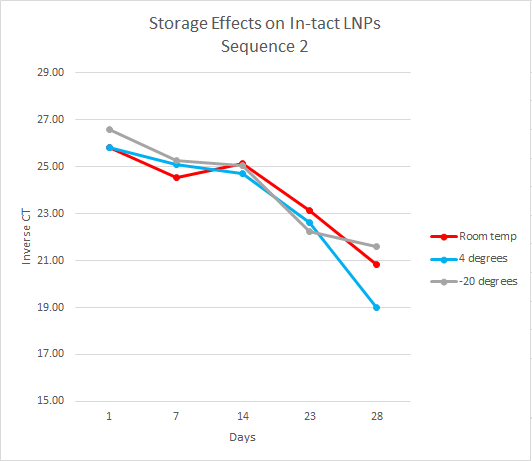
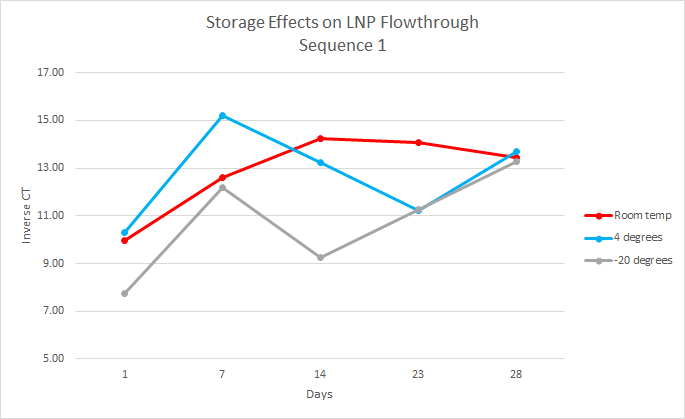
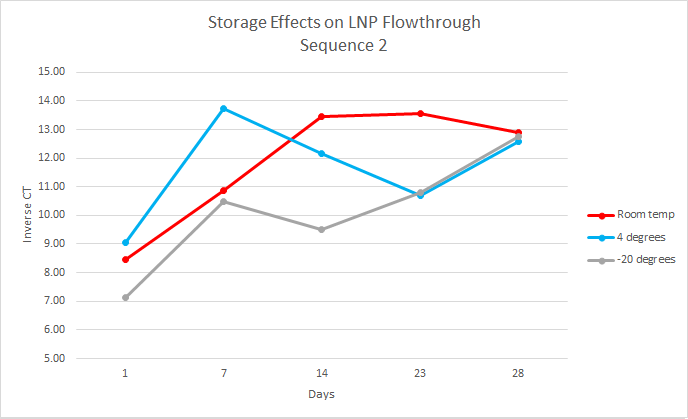
For this experiment, the first step, which had already been done, was to load the LNPs with DNA cargo. This experiment used two DNA sequences, sequence one and sequence two, both 35 base pairs long. The samples were organized by temperature, days, and replicates. At each time point, the samples were separated into intact and flow-through by using 10kDa filters. Over the course of 28 days, the samples were analyzed every week by performing qPCR on the intact and flow-through.
PCR Data Analysis
After running qPCR we exported the data. We then calculated the mean of the technical and biological replicates. This value is the CT value however it is inversely related so to interpret the results easily we took the inverse CT by subtracting the CT value from 40 ( the amount of qPCR cycles). Using this data, we graphed the inverse CT over time for each condition.
Challenges and Troubleshooting
During the first week of the experiment we ran into obstacles. After we had completed the first qPCR our initial samples did not amplify. However the negative control (water) did amplify. After troubleshooting we hypothesized that the phosphate buffer solution was inhibiting the qPCR reaction. To solve this problem the interns dilute the samples 1:10 with water.
Results
We found that over the 28 days the LNPs in the -20 degrees Celsius storage condition had the least amount of leakage therefore it is the most stable storage condition in the experiment.
Mouse Lung Coinfection, Influenza Virus A(flu) and Streptococcus Pneumoniae (strep)
Context
This project investigates the underlying mechanisms of more severe influenza A virus disease in aged individuals. Lungs samples were collected from aged (88 weeks) and young (17 weeks) mice infected with influenza A virus and lung tissue harvested at multiple timepoints for expression array and viral load measurement.
Methods
For this experiment, the RNA from the mouse lungs was isolated in order to synthesize cDNA. Then the cDNA was used to perform qPCR for flu, strep, and both for both the aged (88 weeks) and young mice (17 weeks) at each time point (4 days-18 days).
PCR Data Analysis
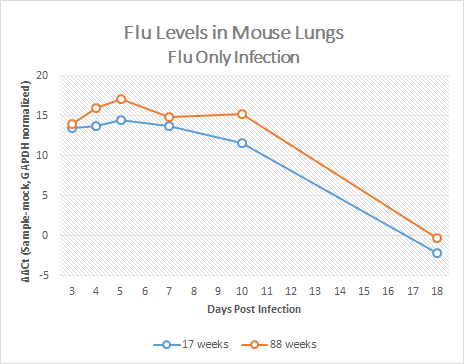
After running qPCR we exported the data. Then we calculated the mean of the technical and biological replicates. After, we then normalized to GAPDH and took the ΔΔCT by subtracting the mock value. Using this data, we graphed the ΔΔCT over time for each condition.
Results
With the data collected we concluded the older mice exhibited higher amounts of flu than the younger mice.
A Note From the Interns
We would like to thank our mentors, Suman Kumari, Kelsey Scherler, and Kathie Walters, for making this a fulfilling summer. We would also like to thank Claudia Ludwig, Sarah Clemente, and the SEE program for offering this experience.