| n.genes | n.arrays | score | residual | profile | network | motif1 | motif2 | motif.posns | orf.names | short.names | long.names |
| 10 | 42 | 324 | -7.351 | 0.135 |  |  | 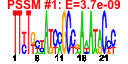 |  |  | b1888 b1887 b1072 b1075 b1076 b1077 b1078 b1079 b1080 b1081 b1922 b1923 b1924 b1937 b1938 b1940 b1941 b1944 b1947 b1949 b1950 b1925 b1926 b1921 b1890 b1889 b1223 b1883 b1884 b1073 b1074 b1082 b1083 b1071 b1070 b1939 b1942 b1943 b1945 b1946 b1948 b1194 | cheA cheW flgA flgD flgE flgF flgG flgH flgI flgJ fliA fliC fliD fliE fliF fliH fliI fliL fliO fliQ fliR fliS fliT fliZ motA motB narK cheB cheR flgB flgC flgK flgL flgM flgN fliG fliJ fliK fliM fliN fliP ycgR | fused chemotactic sensory histidine kinase in two-component regulatory system with CheB and CheY: sensory histidine kinase/signal sensing protein | purine-binding chemotaxis protein | assembly protein for flagellar basal-body periplasmic P ring | flagellar hook assembly protein | flagellar hook protein | flagellar component of cell-proximal portion of basal-body rod | flagellar component of cell-distal portion of basal-body rod | flagellar protein of basal-body outer-membrane L ring | predicted flagellar basal body protein | muramidase | RNA polymerase, sigma 28 (sigma F) factor | flagellar filament structural protein (flagellin) | flagellar filament capping protein | flagellar basal-body component | flagellar basal-body MS-ring and collar protein | flagellar biosynthesis protein | flagellum-specific ATP synthase | flagellar export pore protein | flagellar protein potentiates polymerization | predicted chaperone | predicted regulator of FliA activity | proton conductor component of flagella motor | protein that enables flagellar motor rotation | nitrate/nitrite transporter | fused chemotaxis regulator: protein-glutamate methylesterase in two-component regulatory system with CheA | chemotaxis regulator, protein-glutamate methyltransferase | flagellar hook-filament junction protein 1 | flagellar hook-filament junction protein | anti-sigma factor for FliA (sigma 28) | export chaperone for FlgK and FlgL | flagellar motor switching and energizing component | flagellar protein | flagellar hook-length control protein | protein involved in flagellar function |
| 64 | 24 | 337 | -4.295 | 0.381 |  |  | 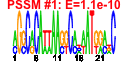 | 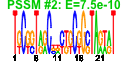 |  | b3198 b3197 b3034 b3030 b3204 b2892 b3202 b3035 b3201 b3205 b3200 b3031 b3191 b3192 b3193 b3196 b3199 b3032 b2893 b2895 b2894 b3033 b3194 b3195 | kdsC kdsD nudF parE ptsN recJ rpoN tolC lptB yhbJ lptA yqiA yrbB yrbC yrbD yrbG yrbK cpdA dsbC fldB xerD yqiB yrbE yrbF | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase | D-arabinose 5-phosphate isomerase | ADP-ribose pyrophosphatase | DNA topoisomerase IV, subunit B | sugar-specific enzyme IIA component of PTS | ssDNA exonuclease, 5' --> 3'-specific | RNA polymerase, sigma 54 (sigma N) factor | transport channel | predicted transporter subunit: ATP-binding component of ABC superfamily | predicted protein with nucleoside triphosphate hydrolase domain | predicted transporter subunit: periplasmic-binding component of ABC superfamily | predicted esterase | predicted protein | predicted ABC-type organic solvent transporter | predicted calcium/sodium:proton antiporter | conserved protein | cyclic 3',5'-adenosine monophosphate phosphodiesterase | protein disulfide isomerase II | flavodoxin 2 | site-specific tyrosine recombinase | predicted dehydrogenase | predicted toluene transporter subunit: membrane component of ABC superfamily | predicted toluene transporter subunit: ATP-binding component of ABC superfamily |
| 104 | 24 | 329 | -4.130 | 0.438 |  |  | 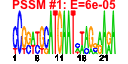 | 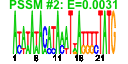 |  | b3323 b3324 b3325 b3326 b3327 b3328 b3329 b3330 b3331 b3332 b3333 b3334 b3335 b3580 b3322 b3582 b3575 b3576 b3577 b3578 b3579 b3678 b3679 b3680 | gspA gspC gspD gspE gspF gspG gspH gspI gspJ gspK gspL gspM gspO lyx gspB sgbU yiaK yiaL yiaM yiaN yiaO yidJ yidK yidL | general secretory pathway component, cryptic | pseudopilin, cryptic, general secretion pathway | predicted general secretory pathway component, cryptic | bifunctional prepilin leader peptidase/ methylase | L-xylulose kinase | part of gsp divergon involved in type II protein secretion | predicted L-xylulose 5-phosphate 3-epimerase | 2,3-diketo-L-gulonate dehydrogenase, NADH-dependent | conserved protein | predicted transporter | predicted sulfatase/phosphatase | predicted DNA-binding transcriptional regulator |
| 147 | 24 | 331 | -3.372 | 0.288 |  |  | 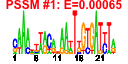 | 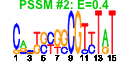 |  | b1746 b1744 b1487 b1486 b1485 b1484 b1483 b1513 b1514 b1515 b1517 b1518 b1511 b1512 b1463 b1388 b1389 b1390 b1391 b1396 b1442 b1443 b1444 b1462 | astD astE ddpA ddpB ddpC ddpD ddpF lsrA lsrC lsrD lsrF lsrG lsrK lsrR nhoA paaA paaB paaC paaD paaI ydcU ydcV ydcW yddH | succinylglutamic semialdehyde dehydrogenase | succinylglutamate desuccinylase | D-ala-D-a la transporter subunit | D-ala-D-ala transporter subunit | fused AI2 transporter subunits of ABC superfamily: ATP-binding components | AI2 transporter | putative autoinducer-2 (AI-2) aldolase | autoinducer-2 (AI-2) modifying protein LsrG | autoinducer-2 (AI-2) kinase | lsr operon transcriptional repressor | N-hydroxyarylamine O-acetyltransferase | predicted multicomponent oxygenase/reductase subunit for phenylacetic acid degradation | predicted thioesterase | predicted spermidine/putrescine transporter subunit | medium chain aldehyde dehydrogenase | conserved protein |
| 144 | 24 | 339 | -3.051 | 0.288 | 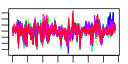 |  | 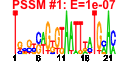 | 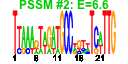 |  | b1337 b1339 b1336 b1759 b1335 b1396 b1692 b1802 b1410 b1411 b1758 b1338 b1592 b1653 b1392 b1393 b1394 b1395 b1397 b1398 b1358 b1803 b1834 b1408 | abgB abgR abgT nudG ogt paaI ydiB yeaW ynbC ynbD ynjF abgA clcB lhr paaE paaF paaG paaH paaJ paaK ydaT yeaX yebT ynbA | predicted peptidase, aminobenzoyl-glutamate utilization protein | predicted DNA-binding transcriptional regulator | predicted cryptic aminobenzoyl-glutamate transporter | pyrimidine (deoxy)nucleoside triphosphate pyrophosphohydrolase | O-6-alkylguanine-DNA:cysteine-protein methyltransferase | predicted thioesterase | quinate/shikimate 5-dehydrogenase, NAD(P)-binding | predicted 2Fe-2S cluster-containing protein | predicted hydrolase | predicted phosphatase, inner membrane protein | predicted phosphatidyl transferase, inner membrane protein | predicted voltage-gated chloride channel | predicted ATP-dependent helicase | predicted multicomponent oxygenase/reductase subunit for phenylacetic acid degradation | enoyl-CoA hydratase-isomerase | acyl-CoA hydratase | 3-hydroxybutyryl-CoA dehydrogenase | predicted beta-ketoadipyl CoA thiolase | phenylacetyl-CoA ligase | Rac prophage; predicted protein | predicted oxidoreductase | conserved protein | predicted inner membrane protein |
| 110 | 33 | 330 | -2.566 | 0.374 | 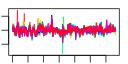 |  | 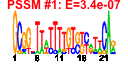 | 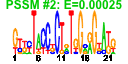 |  | b2198 b1487 b1486 b1485 b1484 b1483 b0873 b0872 b1473 b1412 b2201 b2200 b2199 b2197 b2196 b2195 b2194 b1474 b1475 b1476 b1413 b2206 b2203 b2202 b2207 b2205 b2204 b1227 b1226 b1521 b0992 b1406 b1674 | ccmD ddpA ddpB ddpC ddpD ddpF hcp hcr yddG azoR ccmA ccmB ccmC ccmE ccmF ccmG ccmH fdnG fdnH fdnI hrpA napA napB napC napD napG napH narI narJ uxaB yccM ydbC ydhY | cytochrome c biogenesis protein | D-ala-D-a la transporter subunit | D-ala-D-ala transporter subunit | hybrid-cluster [4Fe-2S-2O] protein in anaerobic terminal reductases | HCP oxidoreductase, NADH-dependent | predicted methyl viologen efflux pump | NADH-azoreductase, FMN-dependent | heme exporter subunit | periplasmic heme chaperone | heme lyase, CcmF subunit | periplasmic thioredoxin of cytochrome c-type biogenesis | heme lyase, CcmH subunit | formate dehydrogenase-N, alpha subunit, nitrate-inducible | formate dehydrogenase-N, Fe-S (beta) subunit, nitrate-inducible | formate dehydrogenase-N, cytochrome B556 (gamma) subunit, nitrate-inducible | ATP-dependent helicase | nitrate reductase, periplasmic, large subunit | nitrate reductase, small, cytochrome C550 subunit, periplasmic | nitrate reductase, cytochrome c-type, periplasmic | assembly protein for periplasmic nitrate reductase | ferredoxin-type protein essential for electron transfer from ubiquinol to periplasmic nitrate reductase (NapAB) | nitrate reductase 1, gamma (cytochrome b(NR)) subunit | molybdenum-cofactor-assembly chaperone subunit (delta subunit) of nitrate reductase 1 | altronate oxidoreductase, NAD-dependent | predicted 4Fe-4S membrane protein | predicted oxidoreductase, NAD(P)-binding | predicted 4Fe-4S ferridoxin-type protein |
| 28 | 27 | 332 | -2.389 | 0.306 |  |  | 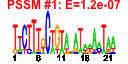 | 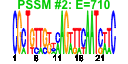 |  | b2049 b2052 b2053 b1377 b2162 b0936 b0933 b0934 b0935 b0937 b2057 b2054 b2050 b2062 b2061 b2060 b0284 b1490 b1690 b1801 b1803 b1862 b2166 b2161 b2163 b1489 b0823 | cpsB fcl gmd ompN rihB ssuA ssuB ssuC ssuD ssuE wcaC wcaF wcaI wza wzb wzc yagR yddV ydiM yeaV yeaX yebB yeiC nupX yeiL dos ybiW | mannose-1-phosphate guanyltransferase | bifunctional GDP-fucose synthetase: GDP-4-dehydro-6-deoxy-D-mannose epimerase/ GDP-4-dehydro-6-L-deoxygalactose reductase | GDP-D-mannose dehydratase, NAD(P)-binding | outer membrane pore protein N, non-specific | ribonucleoside hydrolase 2 | alkanesulfonate transporter subunit | alkanesulfonate monooxygenase, FMNH(2)-dependent | NAD(P)H-dependent FMN reductase | predicted glycosyl transferase | predicted acyl transferase | lipoprotein required for capsular polysaccharide translocation through the outer membrane | protein-tyrosine phosphatase | protein-tyrosine kinase | predicted oxidoreductase with molybdenum-binding domain | predicted diguanylate cyclase | predicted transporter | predicted oxidoreductase | predicted protein | predicted kinase | predicted nucleoside transporter | DNA-binding transcriptional activator of stationary phase nitrogen survival | cAMP phosphodiesterase, heme-regulated | predicted pyruvate formate lyase |
| 60 | 37 | 348 | -2.263 | 0.338 | 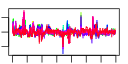 |  | 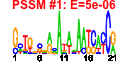 |  |  | b0979 b0978 b0273 b1217 b1040 b1039 b1038 b0575 b0574 b0572 b0573 b0854 b0855 b0856 b0857 b1296 b2103 b2104 b1260 b1261 b1262 b1263 b1264 b1265 b0600 b0980 b1037 b0571 b0570 b0972 b0973 b0974 b0975 b0976 b0977 b1267 b1266 | appB appC argF chaB csgD csgE csgF cusA cusB cusC cusF potF potG potH potI puuP thiD thiM trpA trpB trpC trpD trpE trpL ybdL appA csgG cusR cusS hyaA hyaB hyaC hyaD hyaE hyaF yciO trpH | cytochrome bd-II oxidase, subunit II | cytochrome bd-II oxidase, subunit I | CP4-6 prophage; ornithine carbamoyltransferase 2, chain F | predicted cation regulator | DNA-binding transcriptional activator in two-component regulatory system | predicted transport protein | predicted transport protein | copper/silver efflux system, membrane component | copper/silver efflux system, membrane fusion protein | copper/silver efflux system, outer membrane component | periplasmic copper-binding protein | putrescine transporter subunit: periplasmic-binding component of ABC superfamily | putrescine transporter subunit: ATP-binding component of ABC superfamily | putrescine transporter subunit: membrane component of ABC superfamily | putrescine importer | bifunctional hydroxy-methylpyrimidine kinase/ hydroxy-phosphomethylpyrimidine kinase | hydoxyethylthiazole kinase | tryptophan synthase, alpha subunit | tryptophan synthase, beta subunit | fused indole-3-glycerolphosphate synthetase/N-(5-phosphoribosyl)anthranilate isomerase | fused glutamine amidotransferase (component II) of anthranilate synthase/anthranilate phosphoribosyl transferase | component I of anthranilate synthase | trp operon leader peptide | methionine aminotransferase, PLP-dependent | phosphoanhydride phosphorylase | outer membrane lipoprotein | DNA-binding response regulator in two-component regulatory system with CusS | sensory histidine kinase in two-component regulatory system with CusR, senses copper ions | hydrogenase 1, small subunit | hydrogenase 1, large subunit | hydrogenase 1, b-type cytochrome subunit | protein involved in processing of HyaA and HyaB proteins | protein involved in nickel incorporation into hydrogenase-1 proteins | conserved protein |
| 23 | 58 | 337 | -2.208 | 0.324 | 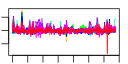 |  | 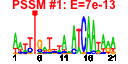 | 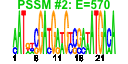 |  | b2957 b3368 b0621 b0894 b0895 b0896 b0972 b0973 b0974 b0975 b0976 b0977 b2725 b2724 b2723 b2722 b2721 b2720 b2719 b2718 b2717 b2726 b2728 b2729 b2730 b0344 b2208 b1224 b1225 b3476 b3477 b3478 b3479 b3480 b3365 b3366 b4070 b4071 b4072 b1256 b0919 b1541 b1750 b1906 b1970 b2110 b2111 b2579 b3158 b3159 b4380 b1587 b1588 b1589 b1593 b2727 b1223 b3367 | ansB cysG dcuC dmsA dmsB dmsC hyaA hyaB hyaC hyaD hyaE hyaF hycA hycB hycC hycD hycE hycF hycG hycH hycI hypA hypC hypD hypE lacZ napF narG narH nikA nikB nikC nikD nikE nirB nirD nrfA nrfB nrfC ompW ycbJ ydfZ ydjX yecH yedX yehC yehD yfiD yhbU yhbV yjjI ynfE ynfF ynfG ynfK hypB narK nirC | periplasmic L-asparaginase II | fused siroheme synthase 1,3-dimethyluroporphyriongen III dehydrogenase and siroheme ferrochelatase/uroporphyrinogen methyltransferase | anaerobic C4-dicarboxylate transport | dimethyl sulfoxide reductase, anaerobic, subunit A | dimethyl sulfoxide reductase, anaerobic, subunit B | dimethyl sulfoxide reductase, anaerobic, subunit C | hydrogenase 1, small subunit | hydrogenase 1, large subunit | hydrogenase 1, b-type cytochrome subunit | protein involved in processing of HyaA and HyaB proteins | protein involved in nickel incorporation into hydrogenase-1 proteins | regulator of the transcriptional regulator FhlA | hydrogenase 3, Fe-S subunit | hydrogenase 3, membrane subunit | hydrogenase 3, large subunit | formate hydrogenlyase complex iron-sulfur protein | hydrogenase 3 and formate hydrogenase complex, HycG subunit | protein required for maturation of hydrogenase 3 | protease involved in processing C-terminal end of HycE | protein involved in nickel insertion into hydrogenases 3 | protein required for maturation of hydrogenases 1 and 3 | protein required for maturation of hydrogenases | carbamoyl phosphate phosphatase, hydrogenase 3 maturation protein | beta-D-galactosidase | ferredoxin-type protein, predicted role in electron transfer to periplasmic nitrate reductase (NapA) | nitrate reductase 1, alpha subunit | nitrate reductase 1, beta (Fe-S) subunit | nickel transporter subunit | nitrite reductase, large subunit, NAD(P)H-binding | nitrite reductase, NAD(P)H-binding, small subunit | nitrite reductase, formate-dependent, cytochrome | nitrite reductase, formate-dependent, penta-heme cytochrome c | formate-dependent nitrite reductase, 4Fe4S subunit | outer membrane protein W | conserved protein | predicted inner membrane protein | predicted protein | predicted periplasmic pilin chaperone | predicted fimbrial-like adhesin protein | pyruvate formate lyase subunit | predicted peptidase (collagenase-like) | predicted protease | oxidoreductase subunit | oxidoreductase, Fe-S subunit | predicted dethiobiotin synthetase | GTP hydrolase involved in nickel liganding into hydrogenases | nitrate/nitrite transporter | nitrite transporter |
| 71 | 27 | 338 | -2.173 | 0.297 |  |  | 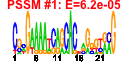 | 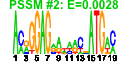 |  | b1000 b0999 b0484 b2097 b2661 b2662 b0450 b1051 b1482 b0485 b0486 b0753 b1836 b2080 b2659 b3003 b1952 b1732 b1646 b1188 b1257 b1258 b1259 b1784 b1955 b1678 b1953 | cbpA cbpM copA fbaB gabD gabT glnK msyB osmC ybaS ybaT ybgS yebV yegP csiD yghA dsrB katE sodC ycgB yciE yciF yciG yeaH yedP ynhG yodD | curved DNA-binding protein, DnaJ homologue that functions as a co-chaperone of DnaK | modulator of CbpA co-chaperone | copper transporter | fructose-bisphosphate aldolase class I | succinate-semialdehyde dehydrogenase I, NADP-dependent | 4-aminobutyrate aminotransferase, PLP-dependent | nitrogen assimilation regulatory protein for GlnL, GlnE, and AmtB | predicted protein | osmotically inducible, stress-inducible membrane protein | predicted glutaminase | predicted transporter | conserved protein | predicted glutathionylspermidine synthase, with NAD(P)-binding Rossmann-fold domain | hydroperoxidase HPII(III) (catalase) | superoxide dismutase, Cu, Zn |
| 107 | 49 | 333 | -2.115 | 0.259 |  |  | 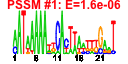 |  |  | b3960 b0750 b0751 b2818 b3959 b3958 b3359 b0273 b3172 b4254 b0754 b0112 b0860 b0336 b0572 b0573 b2750 b2752 b3213 b0078 b0077 b0074 b0073 b0072 b0071 b0075 b3454 b3455 b3457 b3460 b3458 b3456 b2838 b4024 b4013 b3008 b3829 b3941 b3661 b2476 b2557 b4245 b4244 b2913 b3994 b3991 b3990 b4407 b3523 | argH nadA pnuC argA argB argC argD argF argG argI aroG aroP artJ codB cusC cusF cysC cysD gltD ilvH ilvI leuA leuB leuC leuD leuL livF livG livH livJ livK livM lysA lysC metA metC metE metF nlpA purC purL pyrB pyrI serA thiC thiG thiH thiS yhjE | argininosuccinate lyase | quinolinate synthase, subunit A | predicted nicotinamide mononucleotide transporter | fused acetylglutamate kinase homolog (inactive)/amino acid N-acetyltransferase | acetylglutamate kinase | N-acetyl-gamma-glutamylphosphate reductase, NAD(P)-binding | bifunctional acetylornithine aminotransferase/ succinyldiaminopimelate aminotransferase | CP4-6 prophage; ornithine carbamoyltransferase 2, chain F | argininosuccinate synthetase | ornithine carbamoyltransferase 1 | 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase, phenylalanine repressible | aromatic amino acid transporter | arginine transporter subunit | cytosine transporter | copper/silver efflux system, outer membrane component | periplasmic copper-binding protein | adenosine 5'-phosphosulfate kinase | sulfate adenylyltransferase, subunit 2 | glutamate synthase, 4Fe-4S protein, small subunit | acetolactate synthase III, thiamin-dependent, small subunit | acetolactate synthase III, large subunit | 2-isopropylmalate synthase | 3-isopropylmalate dehydrogenase | 3-isopropylmalate isomerase subunit, dehydratase component | 3-isopropylmalate isomerase subunit | leu operon leader peptide | leucine/isoleucine/valine transporter subunit | leucine transporter subunit | diaminopimelate decarboxylase, PLP-binding | aspartokinase III | homoserine O-transsuccinylase | cystathionine beta-lyase, PLP-dependent | 5-methyltetrahydropteroyltriglutamate- homocysteine S-methyltransferase | 5,10-methylenetetrahydrofolate reductase | cytoplasmic membrane lipoprotein-28 | phosphoribosylaminoimidazole-succinocarboxamide synthetase | phosphoribosylformyl-glycineamide synthetase | aspartate carbamoyltransferase, catalytic subunit | aspartate carbamoyltransferase, regulatory subunit | D-3-phosphoglycerate dehydrogenase | thiamin (pyrimidine moiety) biosynthesis protein | thiamin biosynthesis ThiGH complex subunit | sulphur carrier protein | predicted transporter |
| 131 | 42 | 331 | -2.040 | 0.368 |  |  |  |  |  | b3206 b3203 b3389 b3390 b3387 b3809 b4207 b3850 b3198 b3197 b0168 b3972 b2830 b4162 b2908 b3847 b4160 b3204 b2829 b0196 b3619 b3620 b3202 b4161 b2620 b3849 b2907 b2906 b3811 b2909 b3201 b3205 b3200 b3345 b3346 b4558 b3810 b3191 b3192 b3193 b3196 b3199 | npr hpf aroB aroK dam dapF fklB hemG kdsC kdsD map murB nudH orn pepP pepQ psd ptsN ptsP rcsF rfaD rfaF rpoN rsgA smpB trkH ubiH visC xerC ygfB lptB yhbJ lptA yheN yheO yifL yigA yrbB yrbC yrbD yrbG yrbK | phosphohistidinoprotein-hexose phosphotransferase component of N-regulated PTS system (Npr) | predicted ribosome-associated, sigma 54 modulation protein | 3-dehydroquinate synthase | shikimate kinase I | DNA adenine methylase | diaminopimelate epimerase | FKBP-type peptidyl-prolyl cis-trans isomerase (rotamase) | protoporphyrin oxidase, flavoprotein | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase | D-arabinose 5-phosphate isomerase | methionine aminopeptidase | UDP-N-acetylenolpyruvoylglucosamine reductase, FAD-binding | nucleotide hydrolase | oligoribonuclease | proline aminopeptidase P II | proline dipeptidase | phosphatidylserine decarboxylase | sugar-specific enzyme IIA component of PTS | fused PTS enzyme: PEP-protein phosphotransferase (enzyme I)/GAF domain containing protein | predicted outer membrane protein, signal | ADP-L-glycero-D-mannoheptose-6-epimerase, NAD(P)-binding | ADP-heptose:LPS heptosyltransferase II | RNA polymerase, sigma 54 (sigma N) factor | ribosome small subunit-dependent GTPase A | trans-translation protein | potassium transporter | 2-octaprenyl-6-methoxyphenol hydroxylase, FAD/NAD(P)-binding | predicted oxidoreductase with FAD/NAD(P)-binding domain | site-specific tyrosine recombinase | predicted protein | predicted transporter subunit: ATP-binding component of ABC superfamily | predicted protein with nucleoside triphosphate hydrolase domain | predicted transporter subunit: periplasmic-binding component of ABC superfamily | predicted intracellular sulfur oxidation protein | predicted DNA-binding transcriptional regulator | predicted lipoprotein | conserved protein | predicted ABC-type organic solvent transporter | predicted calcium/sodium:proton antiporter |
| 135 | 39 | 335 | -2.035 | 0.302 |  |  | 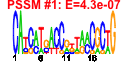 | 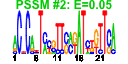 |  | b1338 b1592 b0997 b1408 b2049 b1556 b1581 b1580 b0532 b1547 b1372 b0996 b0998 b0995 b1873 b1872 b2050 b2047 b1202 b1407 b1509 b1543 b1553 b1554 b1555 b1559 b1560 b1669 b1670 b1673 b1672 b1671 b1701 b1698 b1999 b2002 b2005 b2121 b1409 | abgA clcB torA ynbA cpsB essQ rspA rspB sfmD ydfN stfR torC torD torR torY torZ wcaI wcaJ ycgV ydbD ydeU ydfP ydfQ ydfR ydfT ydfU ydhT ydhU ydhV ydhW ydhX fadK ydiR yeeP yeeS yeeV yehP ynbB | predicted peptidase, aminobenzoyl-glutamate utilization protein | predicted voltage-gated chloride channel | trimethylamine N-oxide (TMAO) reductase I, catalytic subunit | predicted inner membrane protein | mannose-1-phosphate guanyltransferase | Qin prophage; predicted S lysis protein | predicted dehydratase | predicted oxidoreductase, Zn-dependent and NAD(P)-binding | predicted outer membrane export usher protein | Qin prophage; predicted side tail fibre assembly protein | Rac prophage; predicted tail fiber protein | trimethylamine N-oxide (TMAO) reductase I, cytochrome c-type subunit | chaperone involved in maturation of TorA subunit of trimethylamine N-oxide reductase system I | DNA-binding response regulator in two-component regulatory system with TorS | TMAO reductase III (TorYZ), cytochrome c-type subunit | trimethylamine N-oxide reductase system III, catalytic subunit | predicted glycosyl transferase | predicted UDP-glucose lipid carrier transferase | predicted adhesin | predicted protein | conserved protein | Qin prophage; conserved protein | Qin prophage; predicted lysozyme | Qin prophage; predicted protein | Qin prophage; predicted antitermination protein Q | predicted cytochrome | predicted oxidoreductase | predicted 4Fe-4S ferridoxin-type protein | short chain acyl-CoA synthetase, anaerobic | predicted electron transfer flavoprotein, FAD-binding | CP4-44 prophage; predicted DNA repair protein | CP4-44 prophage; toxin of the YeeV-YeeU toxin-antitoxin system | predicted CDP-diglyceride synthase |
| 56 | 37 | 341 | -2.013 | 0.259 |  |  | 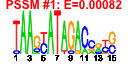 | 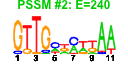 |  | b1478 b0980 b1747 b1745 b1748 b1732 b1988 b1896 b1646 b1006 b1007 b1008 b1011 b1012 b1188 b1257 b1258 b1259 b1440 b1441 b1784 b1932 b1955 b0836 b1195 b1678 b1953 b2135 b2137 b1488 b1009 b1684 b1683 b1681 b1679 b1680 b1010 | adhP appA astA astB astC katE nac otsA sodC rutG rutF rutE rutB rutA ycgB yciE yciF yciG ydcS ydcT yeaH yedL yedP bssR ymgE ynhG yodD yohC yohF ddpX rutD sufA sufB sufD sufE sufS rutC | ethanol-active dehydrogenase/acetaldehyde-active reductase | phosphoanhydride phosphorylase | arginine succinyltransferase | succinylarginine dihydrolase | succinylornithine transaminase, PLP-dependent | hydroperoxidase HPII(III) (catalase) | DNA-binding transcriptional dual regulator of nitrogen assimilation | trehalose-6-phosphate synthase | superoxide dismutase, Cu, Zn | predicted transporter | predicted oxidoreductase, flavin:NADH component | predicted oxidoreductase | predicted enzyme | predicted monooxygenase | conserved protein | predicted protein | predicted spermidine/putrescine transporter subunit | predicted acyltransferase | predicted inner membrane protein | predicted oxidoreductase with NAD(P)-binding Rossmann-fold domain | D-ala-D-ala dipeptidase, Zn-dependent | predicted hydrolase | Fe-S cluster assembly protein | component of SufBCD complex | sulfur acceptor protein | selenocysteine lyase, PLP-dependent |
| 118 | 32 | 325 | -1.822 | 0.309 |  |  | 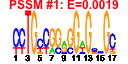 |  |  | b1345 b1311 b1433 b2245 b2437 b2138 b2547 b1385 b1387 b1319 b1158 b1386 b1617 b1615 b1310 b1312 b1313 b1314 b1315 b1316 b1317 b1346 b1472 b1690 b2246 b2247 b2248 b2355 b2544 b2545 b2546 b2549 | intR ycjO ydcO yfaU eutR yphE feaB maoC ompG pinE tynA uidA uidC ycjN ycjP ycjQ ycjR ycjS ycjT ycjU ydaQ yddL ydiM yfaV yfaW yfaX yfdL yphB yphC yphD yphG | Rac prophage; integrase | predicted sugar transporter subunit: membrane component of ABC superfamily | predicted benzoate transporter | predicted 2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase | predicted DNA-binding transcriptional regulator | fused predicted sugar transporter subunits of ABC superfamily: ATP-binding components | phenylacetaldehyde dehydrogenase | fused aldehyde dehydrogenase/enoyl-CoA hydratase | outer membrane porin | e14 prophage; site-specific DNA recombinase | tyramine oxidase, copper-requiring | beta-D-glucuronidase | predicted outer membrane porin protein | predicted sugar transporter subunit: periplasmic-binding component of ABC superfamily | predicted sugar transporter subunit: membrane component of ABC superfamily | predicted oxidoreductase, Zn-dependent and NAD(P)-binding | predicted enzyme | predicted oxidoreductase, NADH-binding | predicted hydrolase | predicted beta-phosphoglucomutase | Rac prophage; conserved protein | predicted transporter | predicted enolase | conserved protein |
| 150 | 33 | 330 | -1.675 | 0.320 |  |  |  |  |  | b1337 b1339 b1336 b1343 b1334 b1345 b4426 b1759 b1335 b1333 b1340 b1341 b1342 b1344 b1414 b1473 b1604 b1639 b1640 b1692 b1751 b1752 b1781 b1802 b1410 b1411 b1525 b1526 b1753 b1754 b1755 b1756 b1758 | abgB abgR abgT dbpA fnr intR nudG ogt uspE ydaL ydaM ydaN ttcA ydcF yddG ydgH ydhA anmK ydiB ydjY ydjZ yeaE yeaW ynbC ynbD yneI yneJ ynjA ynjB ynjC ynjD ynjF | predicted peptidase, aminobenzoyl-glutamate utilization protein | predicted DNA-binding transcriptional regulator | predicted cryptic aminobenzoyl-glutamate transporter | ATP-dependent RNA helicase, specific for 23S rRNA | DNA-binding transcriptional dual regulator, global regulator of anaerobic growth | Rac prophage; integrase | pyrimidine (deoxy)nucleoside triphosphate pyrophosphohydrolase | O-6-alkylguanine-DNA:cysteine-protein methyltransferase | stress-induced protein | conserved protein | predicted diguanylate cyclase, GGDEF domain signalling protein | predicted Zn(II) transporter | predicted C32 tRNA thiolase | predicted methyl viologen efflux pump | predicted protein | predicted lipoprotein | quinate/shikimate 5-dehydrogenase, NAD(P)-binding | conserved inner membrane protein | predicted oxidoreductase | predicted 2Fe-2S cluster-containing protein | predicted hydrolase | predicted phosphatase, inner membrane protein | predicted aldehyde dehydrogenase | fused transporter subunits of ABC superfamily: membrane components | predicted transporter subunit: ATP-binding component of ABC superfamily | predicted phosphatidyl transferase, inner membrane protein |
| 5 | 56 | 332 | -1.646 | 0.386 |  |  | 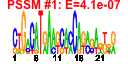 | 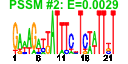 |  | b4169 b3281 b3743 b3287 b4382 b4383 b4381 b4384 b3702 b3701 b3288 b3463 b3462 b3464 b3699 b4174 b4173 b4172 b4372 b4170 b3863 b2677 b2678 b3700 b3628 b3621 b3631 b3627 b3626 b3630 b3632 b3629 b3625 b3624 b3784 b3791 b3787 b3786 b3788 b3789 b3289 b4473 b3284 b3290 b3792 b3785 b3465 b3466 b3644 b4168 b4166 b3096 b3280 b3282 b3283 b4046 | amiB aroE asnC def deoA deoB deoC deoD dnaA dnaN fmt ftsE ftsX ftsY gyrB hflK hflX hfq holD mutL polA proV proW recF rfaB rfaC rfaG rfaI rfaJ rfaP rfaQ rfaS rfaY rfaZ rfe rffA rffD rffE rffG rffH rsmB smf smg trkA wzxE wzzE rsmD yhhL yicC yjeE yjeS yqjB yrdB rimN yrdD zur | N-acetylmuramoyl-l-alanine amidase II | dehydroshikimate reductase, NAD(P)-binding | DNA-binding transcriptional dual regulator | peptide deformylase | thymidine phosphorylase | phosphopentomutase | 2-deoxyribose-5-phosphate aldolase, NAD(P)-linked | purine-nucleoside phosphorylase | chromosomal replication initiator protein DnaA, DNA-binding transcriptional dual regulator | DNA polymerase III, beta subunit | 10-formyltetrahydrofolate:L-methionyl-tRNA(fMet) N-formyltransferase | predicted transporter subunit: ATP-binding component of ABC superfamily | predicted transporter subunit: membrane component of ABC superfamily | fused Signal Recognition Particle (SRP) receptor: membrane binding protein/conserved protein | DNA gyrase, subunit B | modulator for HflB protease specific for phage lambda cII repressor | predicted GTPase | HF-I, host factor for RNA phage Q beta replication | DNA polymerase III, psi subunit | methyl-directed mismatch repair protein | fused DNA polymerase I 5'->3' exonuclease/3'->5' polymerase/3'->5' exonuclease | glycine betaine transporter subunit | gap repair protein | UDP-D-galactose:(glucosyl)lipopolysaccharide-1, 6-D-galactosyltransferase | ADP-heptose:LPS heptosyl transferase I | glucosyltransferase I | UDP-D-galactose:(glucosyl)lipopolysaccharide- alpha-1,3-D-galactosyltransferase | UDP-D-glucose:(galactosyl)lipopolysaccharide glucosyltransferase | kinase that phosphorylates core heptose of lipopolysaccharide | lipopolysaccharide core biosynthesis protein | UDP-GlcNAc:undecaprenylphosphate GlcNAc-1-phosphate transferase | TDP-4-oxo-6-deoxy-D-glucose transaminase | UDP-N-acetyl-D-mannosaminuronic acid dehydrogenase | UDP-N-acetyl glucosamine-2-epimerase | dTDP-glucose 4,6-dehydratase | glucose-1-phosphate thymidylyltransferase | 16S rRNA m5C967 methyltransferase, S-adenosyl-L-methionine-dependent | conserved protein | NAD-binding component of TrK potassium transporter | O-antigen translocase | Entobacterial Common Antigen (ECA) polysaccharide chain length modulation protein | predicted methyltransferase | conserved inner membrane protein | ATPase with strong ADP affinity | predicted Fe-S electron transport protein | predicted ribosome maturation factor | predicted DNA topoisomerase | DNA-binding transcriptional repressor, Zn(II)-binding |
| 31 | 35 | 331 | -1.646 | 0.330 |  |  | 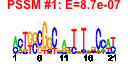 |  |  | b2610 b3844 b2748 b3648 b3168 b2828 b3189 b3967 b3169 b3821 b4375 b3167 b3783 b4373 b3704 b3643 b3703 b4371 b3228 b3838 b2827 b3166 b3843 b3833 b3170 b3705 b4557 b4374 b3194 b3195 b3699 b4372 b3700 b2892 b3644 | ffh fre ftsB gmk infB lgt murA murI nusA pldA prfC rbfA rho rimI rnpA rph rpmH rsmC sspB tatB thyA truB ubiD ubiE yhbC yidC yidD yjjG yrbE yrbF gyrB holD recF recJ yicC | Signal Recognition Particle (SRP) component with 4.5S RNA (ffs) | flavin reductase | cell division protein | guanylate kinase | fused protein chain initiation factor 2, IF2: membrane protein/conserved protein | phosphatidylglycerol-prolipoprotein diacylglyceryl transferase | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | glutamate racemase | transcription termination/antitermination L factor | outer membrane phospholipase A | peptide chain release factor RF-3 | 30s ribosome binding factor | transcription termination factor | acetylase for 30S ribosomal subunit protein S18 | protein C5 component of RNase P | defective ribonuclease PH | 50S ribosomal subunit protein L34 | 16S rRNA m2G1207 methylase | ClpXP protease specificity-enhancing factor | TatABCE protein translocation system subunit | thymidylate synthetase | tRNA pseudouridine synthase | 3-octaprenyl-4-hydroxybenzoate decarboxylase | bifunctional 2-octaprenyl-6-methoxy-1,4-benzoquinone methylase/ S-adenosylmethionine:2-DMK methyltransferase | conserved protein | cytoplasmic insertase into membrane protein, Sec system | predicted protein | dUMP phosphatase | predicted toluene transporter subunit: membrane component of ABC superfamily | predicted toluene transporter subunit: ATP-binding component of ABC superfamily | DNA gyrase, subunit B | DNA polymerase III, psi subunit | gap repair protein | ssDNA exonuclease, 5' --> 3'-specific |
| 125 | 38 | 339 | -1.606 | 0.336 |  |  | 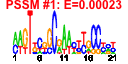 |  |  | b0168 b0637 b2951 b2953 b3639 b3640 b4147 b2610 b2963 b3251 b3189 b2961 b2891 b4375 b0171 b3783 b3704 b4203 b3301 b3294 b3312 b3302 b3703 b3299 b3295 b3230 b3297 b3298 b3311 b3229 b4414 b3641 b2960 b2959 b2952 b2962 b2939 b3190 | map ybeB yggS yggU dfp dut efp ffh mltC mreB murA mutY prfB prfC pyrH rho rnpA rplI rplO rplQ rpmC rpmD rpmH rpmJ rpoA rpsI rpsK rpsM rpsQ sspA slmA trmI yggL yggT yggX yqgB yrbA | methionine aminopeptidase | predicted protein | predicted enzyme | conserved protein | fused 4'-phosphopantothenoylcysteine decarboxylase/phosphopantothenoylcysteine synthetase, FMN-binding | deoxyuridinetriphosphatase | Elongation factor EF-P | Signal Recognition Particle (SRP) component with 4.5S RNA (ffs) | membrane-bound lytic murein transglycosylase C | cell wall structural complex MreBCD, actin-like component MreB | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | adenine DNA glycosylase | peptide chain release factor RF-2 | peptide chain release factor RF-3 | uridylate kinase | transcription termination factor | protein C5 component of RNase P | 50S ribosomal subunit protein L9 | 50S ribosomal subunit protein L15 | 50S ribosomal subunit protein L17 | 50S ribosomal subunit protein L29 | 50S ribosomal subunit protein L30 | 50S ribosomal subunit protein L34 | 50S ribosomal subunit protein L36 | RNA polymerase, alpha subunit | 30S ribosomal subunit protein S9 | 30S ribosomal subunit protein S11 | 30S ribosomal subunit protein S13 | 30S ribosomal subunit protein S17 | stringent starvation protein A | division inhibitor | tRNA (m7G46) methyltransferase, SAM-dependent | predicted inner membrane protein | protein that protects iron-sulfur proteins against oxidative damage | predicted DNA-binding transcriptional regulator |
| 61 | 40 | 333 | -1.584 | 0.330 | 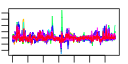 |  | 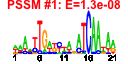 | 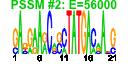 |  | b1474 b2206 b2203 b2207 b2205 b2204 b1227 b1226 b1751 b1757 b2957 b0621 b0894 b0895 b0896 b2728 b0343 b0344 b2208 b1224 b1225 b3476 b3478 b3479 b3365 b3366 b4070 b4071 b4072 b4073 b1256 b1541 b1750 b1906 b2111 b3158 b1587 b1588 b1589 b1593 | fdnG napA napB napD napG napH narI narJ ydjY ynjE ansB dcuC dmsA dmsB dmsC hypC lacY lacZ napF narG narH nikA nikC nikD nirB nirD nrfA nrfB nrfC nrfD ompW ydfZ ydjX yecH yehD yhbU ynfE ynfF ynfG ynfK | formate dehydrogenase-N, alpha subunit, nitrate-inducible | nitrate reductase, periplasmic, large subunit | nitrate reductase, small, cytochrome C550 subunit, periplasmic | assembly protein for periplasmic nitrate reductase | ferredoxin-type protein essential for electron transfer from ubiquinol to periplasmic nitrate reductase (NapAB) | nitrate reductase 1, gamma (cytochrome b(NR)) subunit | molybdenum-cofactor-assembly chaperone subunit (delta subunit) of nitrate reductase 1 | predicted protein | predicted thiosulfate sulfur transferase | periplasmic L-asparaginase II | anaerobic C4-dicarboxylate transport | dimethyl sulfoxide reductase, anaerobic, subunit A | dimethyl sulfoxide reductase, anaerobic, subunit B | dimethyl sulfoxide reductase, anaerobic, subunit C | protein required for maturation of hydrogenases 1 and 3 | lactose/galactose transporter | beta-D-galactosidase | ferredoxin-type protein, predicted role in electron transfer to periplasmic nitrate reductase (NapA) | nitrate reductase 1, alpha subunit | nitrate reductase 1, beta (Fe-S) subunit | nickel transporter subunit | nitrite reductase, large subunit, NAD(P)H-binding | nitrite reductase, NAD(P)H-binding, small subunit | nitrite reductase, formate-dependent, cytochrome | nitrite reductase, formate-dependent, penta-heme cytochrome c | formate-dependent nitrite reductase, 4Fe4S subunit | formate-dependent nitrite reductase, membrane subunit | outer membrane protein W | conserved protein | predicted inner membrane protein | predicted fimbrial-like adhesin protein | predicted peptidase (collagenase-like) | oxidoreductase subunit | oxidoreductase, Fe-S subunit | predicted dethiobiotin synthetase |
| 103 | 45 | 325 | -1.558 | 0.325 | 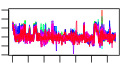 |  | 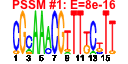 | 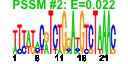 |  | b2818 b3172 b4254 b0754 b0112 b0860 b0336 b3213 b0078 b0077 b3454 b3455 b3457 b3460 b3458 b3456 b4024 b4013 b3008 b3829 b3941 b2476 b4005 b2557 b4245 b4244 b2913 b3523 b0032 b0033 b0337 b2313 b3212 b0198 b0199 b0197 b0523 b2312 b4006 b0522 b2499 b2500 b1849 b1062 b3654 | argA argG argI aroG aroP artJ codB gltD ilvH ilvI livF livG livH livJ livK livM lysC metA metC metE metF purC purD purL pyrB pyrI serA yhjE carA carB codA cvpA gltB metI metN metQ purE purF purH purK purM purN purT pyrC yicE | fused acetylglutamate kinase homolog (inactive)/amino acid N-acetyltransferase | argininosuccinate synthetase | ornithine carbamoyltransferase 1 | 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase, phenylalanine repressible | aromatic amino acid transporter | arginine transporter subunit | cytosine transporter | glutamate synthase, 4Fe-4S protein, small subunit | acetolactate synthase III, thiamin-dependent, small subunit | acetolactate synthase III, large subunit | leucine/isoleucine/valine transporter subunit | leucine transporter subunit | aspartokinase III | homoserine O-transsuccinylase | cystathionine beta-lyase, PLP-dependent | 5-methyltetrahydropteroyltriglutamate- homocysteine S-methyltransferase | 5,10-methylenetetrahydrofolate reductase | phosphoribosylaminoimidazole-succinocarboxamide synthetase | phosphoribosylglycinamide synthetase phosphoribosylamine-glycine ligase | phosphoribosylformyl-glycineamide synthetase | aspartate carbamoyltransferase, catalytic subunit | aspartate carbamoyltransferase, regulatory subunit | D-3-phosphoglycerate dehydrogenase | predicted transporter | carbamoyl phosphate synthetase small subunit, glutamine amidotransferase | carbamoyl-phosphate synthase large subunit | cytosine deaminase | membrane protein required for colicin V production | glutamate synthase, large subunit | DL-methionine transporter subunit | N5-carboxyaminoimidazole ribonucleotide mutase | amidophosphoribosyltransferase | fused IMP cyclohydrolase/phosphoribosylaminoimidazolecarboxamide formyltransferase | N5-carboxyaminoimidazole ribonucleotide synthase | phosphoribosylaminoimidazole synthetase | phosphoribosylglycinamide formyltransferase 1 | phosphoribosylglycinamide formyltransferase 2 | dihydro-orotase |
| 148 | 43 | 342 | -1.557 | 0.324 |  |  |  | 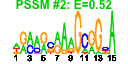 |  | b1094 b0451 b0979 b0978 b2310 b0674 b1597 b1481 b1987 b1218 b1040 b1039 b1038 b2421 b0839 b1095 b1093 b1761 b0655 b2309 b2307 b2306 b2308 b1712 b0912 b0345 b0854 b1089 b4430 b4441 b2104 b1265 b0600 b0801 b1088 b1087 b1287 b1685 b1686 b1687 b0838 b1195 b2135 | acpP amtB appB appC argT asnB asr bdm cbl chaC csgD csgE csgF cysM dacC fabF fabG gdhA gltI hisJ hisM hisP hisQ ihfA ihfB lacI potF rpmF thiM trpL ybdL ybiC yceD yceF yciW ydiH ydiI ydiJ yliJ ymgE yohC | acyl carrier protein (ACP) | ammonium transporter | cytochrome bd-II oxidase, subunit II | cytochrome bd-II oxidase, subunit I | lysine/arginine/ornithine transporter subunit | asparagine synthetase B | acid shock-inducible periplasmic protein | biofilm-dependent modulation protein | DNA-binding transcriptional activator of cysteine biosynthesis | regulatory protein for cation transport | DNA-binding transcriptional activator in two-component regulatory system | predicted transport protein | predicted transport protein | cysteine synthase B (O-acetylserine sulfhydrolase B) | D-alanyl-D-alanine carboxypeptidase (penicillin-binding protein 6a) | 3-oxoacyl-[acyl-carrier-protein] synthase II | 3-oxoacyl-[acyl-carrier-protein] reductase | glutamate dehydrogenase, NADP-specific | glutamate and aspartate transporter subunit | histidine/lysine/arginine/ornithine transporter subunit | integration host factor (IHF), DNA-binding protein, alpha subunit | integration host factor (IHF), DNA-binding protein, beta subunit | DNA-binding transcriptional repressor | putrescine transporter subunit: periplasmic-binding component of ABC superfamily | 50S ribosomal subunit protein L32 | hydoxyethylthiazole kinase | trp operon leader peptide | methionine aminotransferase, PLP-dependent | predicted dehydrogenase | conserved protein | predicted protein | predicted oxidoreductase | predicted FAD-linked oxidoreductase | predicted glutathione S-transferase | predicted inner membrane protein |
| 76 | 39 | 339 | -1.548 | 0.390 |  |  |  | 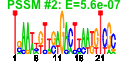 |  | b3389 b3390 b3387 b3639 b3640 b3997 b3850 b4390 b4162 b3847 b4160 b4389 b3619 b3620 b3987 b3649 b4161 b4388 b3229 b3836 b3839 b3849 b3641 b3835 b3834 b3848 b4387 b3648 b4386 b3034 b3030 b3228 b3838 b3035 b3833 b2896 b2897 b2898 b3031 | aroB aroK dam dfp dut hemE hemG nadR orn pepQ psd radA rfaD rfaF rpoB rpoZ rsgA serB sspA tatA tatC trkH slmA ubiB yigP yigZ ytjB gmk lplA nudF parE sspB tatB tolC ubiE ygfX ygfY ygfZ yqiA | 3-dehydroquinate synthase | shikimate kinase I | DNA adenine methylase | fused 4'-phosphopantothenoylcysteine decarboxylase/phosphopantothenoylcysteine synthetase, FMN-binding | deoxyuridinetriphosphatase | uroporphyrinogen decarboxylase | protoporphyrin oxidase, flavoprotein | bifunctional DNA-binding transcriptional repressor/ NMN adenylyltransferase | oligoribonuclease | proline dipeptidase | phosphatidylserine decarboxylase | predicted repair protein | ADP-L-glycero-D-mannoheptose-6-epimerase, NAD(P)-binding | ADP-heptose:LPS heptosyltransferase II | RNA polymerase, beta subunit | RNA polymerase, omega subunit | ribosome small subunit-dependent GTPase A | 3-phosphoserine phosphatase | stringent starvation protein A | TatABCE protein translocation system subunit | potassium transporter | division inhibitor | 2-octaprenylphenol hydroxylase | conserved protein | predicted elongation factor | guanylate kinase | lipoate-protein ligase A | ADP-ribose pyrophosphatase | DNA topoisomerase IV, subunit B | ClpXP protease specificity-enhancing factor | transport channel | bifunctional 2-octaprenyl-6-methoxy-1,4-benzoquinone methylase/ S-adenosylmethionine:2-DMK methyltransferase | predicted protein | predicted folate-dependent regulatory protein | predicted esterase |
| 33 | 51 | 329 | -1.532 | 0.403 | 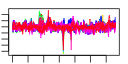 |  | 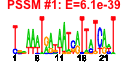 | 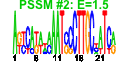 |  | b2155 b1488 b0596 b0595 b0593 b0583 b0594 b0586 b0584 b0592 b0588 b0590 b0589 b0585 b1102 b4367 b0805 b2392 b2675 b2674 b1009 b1684 b1683 b1681 b1679 b1680 b1252 b0591 b0597 b4511 b0803 b0804 b1019 b1018 b1705 b4567 b1452 b2211 b3006 b3005 b4293 b4292 b0150 b0153 b0151 b0152 b2676 b2673 b1682 b3070 b3071 | cirA ddpX entA entB entC entD entE entF fepA fepB fepC fepD fepG fes fhuE fhuF fiu mntH nrdE nrdI rutD sufA sufB sufD sufE sufS tonB entS ybdB ybdZ ybiI ybiX ycdB ycdO ydiE yjjZ yncE yojI exbB exbD fecI fecR fhuA fhuB fhuC fhuD nrdF nrdH sufC yqjH yqjI | ferric iron-catecholate outer membrane transporter | D-ala-D-ala dipeptidase, Zn-dependent | 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase | isochorismatase | isochorismate synthase 1 | phosphopantetheinyltransferase component of enterobactin synthase multienzyme complex | 2,3-dihydroxybenzoate-AMP ligase component of enterobactin synthase multienzyme complex | enterobactin synthase multienzyme complex component, ATP-dependent | iron-enterobactin outer membrane transporter | iron-enterobactin transporter subunit | enterobactin/ferric enterobactin esterase | ferric-rhodotorulic acid outer membrane transporter | ferric iron reductase involved in ferric hydroximate transport | predicted iron outer membrane transporter | manganese/divalent cation transporter | ribonucleoside-diphosphate reductase 2, alpha subunit | protein that stimulates ribonucleotide reduction | predicted hydrolase | Fe-S cluster assembly protein | component of SufBCD complex | sulfur acceptor protein | selenocysteine lyase, PLP-dependent | membrane spanning protein in TonB-ExbB-ExbD complex | predicted transporter | conserved protein | predicted protein | fused predicted multidrug transport subunits of ABC superfamily: membrane component/ATP-binding component | KpLE2 phage-like element; RNA polymerase, sigma 19 factor | KpLE2 phage-like element; transmembrane signal transducer for ferric citrate transport | ferrichrome outer membrane transporter | fused iron-hydroxamate transporter subunits of ABC superfamily: membrane components | iron-hydroxamate transporter subunit | ribonucleoside-diphosphate reductase 2, beta subunit, ferritin-like protein | glutaredoxin-like protein | component of SufBCD complex, ATP-binding component of ABC superfamily | predicted siderophore interacting protein | predicted transcriptional regulator |
| 99 | 41 | 341 | -1.462 | 0.370 | 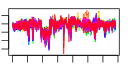 |  | 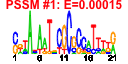 | 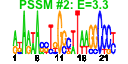 |  | b0185 b0910 b0184 b4410 b4147 b0175 b3187 b0174 b3294 b3312 b3302 b3299 b3295 b3311 b4414 b0176 b3260 b3261 b3978 b3168 b0672 b3169 b3167 b3986 b3186 b3185 b3637 b3936 b3636 b0169 b3296 b3165 b0023 b3065 b3979 b3976 b0436 b3166 b0170 b3170 b4146 | accA cmk dnaE ecnA efp cdsA ispB ispU rplQ rpmC rpmD rpmJ rpoA rpsQ rseP dusB fis glyT infB leuW nusA rbfA rplL rplU rpmA rpmB rpmE rpmG rpsB rpsD rpsO rpsT rpsU thrT thrU tig truB tsf yhbC yjeK | acetyl-CoA carboxylase, carboxytransferase, alpha subunit | cytidylate kinase | DNA polymerase III alpha subunit | entericidin A membrane lipoprotein, antidote entericidin B | Elongation factor EF-P | CDP-diglyceride synthase | octaprenyl diphosphate synthase | undecaprenyl pyrophosphate synthase | 50S ribosomal subunit protein L17 | 50S ribosomal subunit protein L29 | 50S ribosomal subunit protein L30 | 50S ribosomal subunit protein L36 | RNA polymerase, alpha subunit | 30S ribosomal subunit protein S17 | zinc metallopeptidase | tRNA-dihydrouridine synthase B | global DNA-binding transcriptional dual regulator | tRNA-Gly | fused protein chain initiation factor 2, IF2: membrane protein/conserved protein | tRNA-Leu | transcription termination/antitermination L factor | 30s ribosome binding factor | 50S ribosomal subunit protein L7/L12 | 50S ribosomal subunit protein L21 | 50S ribosomal subunit protein L27 | 50S ribosomal subunit protein L28 | 50S ribosomal subunit protein L31 | 50S ribosomal subunit protein L33 | 30S ribosomal subunit protein S2 | 30S ribosomal subunit protein S4 | 30S ribosomal subunit protein S15 | 30S ribosomal subunit protein S20 | 30S ribosomal subunit protein S21 | tRNA-Thr | peptidyl-prolyl cis/trans isomerase (trigger factor) | tRNA pseudouridine synthase | protein chain elongation factor EF-Ts | conserved protein | predicted lysine aminomutase |
| 94 | 39 | 332 | -1.460 | 0.382 |  |  | 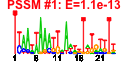 | 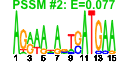 |  | b0587 b0278 b0279 b0719 b0716 b0717 b2108 b2505 b2888 b1455 b0517 b0040 b0297 b0041 b0042 b0043 b0044 b0139 b0331 b0333 b0334 b0335 b0330 b2059 b2058 b2056 b2055 b0045 b0252 b0521 b0824 b2506 b2735 b2736 b2737 b2738 b2739 b0301 b0520 | fepE yagL yagM ybgD ybgO ybgP yehA yfgH ygfU yncH allD caiT eaeH fixA fixB fixC fixX htrE prpB prpC prpD prpE prpR wcaA wcaB wcaD wcaE yaaU yafZ ybcF ybiY yfgI ygbI ygbJ ygbK ygbL ygbM ykgB ylbF | regulator of length of O-antigen component of lipopolysaccharide chains | CP4-6 prophage; DNA-binding protein | CP4-6 prophage; predicted protein | predicted fimbrial-like adhesin protein | predicted assembly protein | predicted outer membrane lipoprotein | predicted transporter | predicted protein | ureidoglycolate dehydrogenase | predicted electron transfer flavoprotein subunit, ETFP adenine nucleotide-binding domain | predicted electron transfer flavoprotein, NAD/FAD-binding domain and ETFP adenine nucleotide-binding domain-like protein | predicted oxidoreductase with FAD/NAD(P)-binding domain | predicted 4Fe-4S ferredoxin-type protein | predicted outer membrane usher protein | 2-methylisocitrate lyase | 2-methylcitrate synthase | 2-methylcitrate dehydratase | predicted propionyl-CoA synthetase with ATPase domain | DNA-binding transcriptional activator | predicted glycosyl transferase | predicted acyl transferase | predicted colanic acid polymerase | CP4-6 prophage; conserved protein | predicted carbamate kinase | predicted pyruvate formate lyase activating enzyme | conserved protein | predicted DNA-binding transcriptional regulator | predicted dehydrogenase, with NAD(P)-binding Rossmann-fold domain | predicted class II aldolase | conserved inner membrane protein |
| 85 | 48 | 335 | -1.352 | 0.406 |  |  | 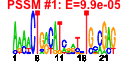 | 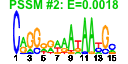 |  | b3809 b4052 b3613 b3729 b3730 b3612 b3779 b3608 b3610 b3805 b3804 b3803 b3802 b3998 b2830 b3780 b3609 b3781 b3611 b3614 b4558 b3810 b3812 b3999 b0050 b3281 b3287 b0055 b3288 b0093 b3997 b4174 b4173 b0054 b0051 b0052 b4389 b3289 b4473 b3284 b0053 b3290 b3848 b4261 b3280 b3282 b3283 b4387 | dapF dnaB envC glmS glmU gpmM gpp gpsA grxC hemC hemD hemX hemY nfi nudH rhlB secB trxA yibN yibQ yifL yigA yigB yjaG apaG aroE def djlA fmt ftsQ hemE hflK hflX imp ksgA pdxA radA rsmB smf smg surA trkA yigZ yjgP yrdB rimN yrdD ytjB | diaminopimelate epimerase | replicative DNA helicase | protease with a role in cell division | L-glutamine:D-fructose-6-phosphate aminotransferase | fused N-acetyl glucosamine-1-phosphate uridyltransferase/glucosamine-1-phosphate acetyl transferase | phosphoglycero mutase III, cofactor-independent | guanosine pentaphosphatase/exopolyphosphatase | glycerol-3-phosphate dehydrogenase (NAD+) | glutaredoxin 3 | hydroxymethylbilane synthase | uroporphyrinogen III synthase | predicted uroporphyrinogen III methylase | predicted protoheme IX synthesis protein | endonuclease V | nucleotide hydrolase | ATP-dependent RNA helicase | protein export chaperone | thioredoxin 1 | predicted rhodanese-related sulfurtransferase | predicted polysaccharide deacetylase | predicted lipoprotein | conserved protein | predicted hydrolase | protein associated with Co2+ and Mg2+ efflux | dehydroshikimate reductase, NAD(P)-binding | peptide deformylase | DnaJ-like protein, membrane anchored | 10-formyltetrahydrofolate:L-methionyl-tRNA(fMet) N-formyltransferase | membrane anchored protein involved in growth of wall at septum | uroporphyrinogen decarboxylase | modulator for HflB protease specific for phage lambda cII repressor | predicted GTPase | exported protein required for envelope biosynthesis and integrity | S-adenosylmethionine-6-N',N'-adenosyl (rRNA) dimethyltransferase | 4-hydroxy-L-threonine phosphate dehydrogenase, NAD-dependent | predicted repair protein | 16S rRNA m5C967 methyltransferase, S-adenosyl-L-methionine-dependent | peptidyl-prolyl cis-trans isomerase (PPIase) | NAD-binding component of TrK potassium transporter | predicted elongation factor | conserved inner membrane protein | predicted ribosome maturation factor | predicted DNA topoisomerase |
| 2 | 37 | 337 | -1.321 | 0.351 |  |  | 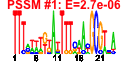 | 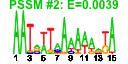 |  | b0649 b0982 b1352 b1348 b1351 b1350 b1349 b1561 b0996 b1873 b1872 b0647 b0648 b0718 b0944 b0941 b0943 b0981 b0968 b0983 b1407 b1472 b1499 b1502 b2072 b2073 b2233 b2270 b2271 b2361 b2362 b2363 b2372 b0984 b0985 b0986 b1506 | djlC etp kilR lar racC recE recT rem torC torY torZ ybeT ybeU ybgQ ycbF ycbT ycbV etk yccX gfcE ydbD yddL ydeO ydeQ yegK yegL yfaL yfbK yfbL yfdR yfdS yfdT yfdV gfcD gfcC gfcB yneL | Hsc56 co-chaperone of HscC | phosphotyrosine-protein phosphatase | Rac prophage; inhibitor of ftsZ, killing protein | Rac prophage; restriction alleviation protein | Rac prophage; predicted protein | Rac prophage; exonuclease VIII, 5' -> 3' specific dsDNA exonuclease | Rac prophage; recombination and repair protein | Qin prophage; predicted protein | trimethylamine N-oxide (TMAO) reductase I, cytochrome c-type subunit | TMAO reductase III (TorYZ), cytochrome c-type subunit | trimethylamine N-oxide reductase system III, catalytic subunit | conserved outer membrane protein | predicted tRNA ligase | predicted outer membrane protein | predicted periplasmic pilini chaperone | predicted fimbrial-like adhesin protein | cryptic autophosphorylating protein tyrosine kinase Etk | predicted acylphosphatase | predicted exopolysaccharide export protein | predicted protein | predicted DNA-binding transcriptional acfivator | conserved protein | adhesin | predicted peptidase | CPS-53 (KpLE1) prophage; conserved protein | CPS-53 (KpLE1) prophage; predicted protein | predicted transporter | predicted outer membrane lipoprotein |
| 27 | 37 | 331 | -1.283 | 0.369 |  |  | 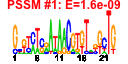 | 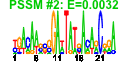 |  | b2068 b2212 b0774 b0777 b0778 b0776 b1042 b1041 b1489 b1385 b1469 b1465 b1466 b1467 b1468 b1838 b0693 b0496 b0788 b0789 b0790 b0964 b1085 b1428 b1536 b1648 b1668 b1985 b2069 b2118 b1454 b1522 b1815 b2213 b1453 b1384 b0808 | alkA alkB bioA bioC bioD bioF csgA csgB dos feaB narU narV narW narY narZ pphA speF ybbP ybhN ybhO ybhP yccT yceQ ydcK ydeI ydhL ydhS yeeO yegD yehI yncG yneF yoaD ada ansP feaR ybiO | 3-methyl-adenine DNA glycosylase II | oxidative demethylase of N1-methyladenine or N3-methylcytosine DNA lesions | 7,8-diaminopelargonic acid synthase, PLP-dependent | predicted methltransferase, enzyme of biotin synthesis | dethiobiotin synthetase | 8-amino-7-oxononanoate synthase | cryptic curlin major subunit | curlin nucleator protein, minor subunit in curli complex | cAMP phosphodiesterase, heme-regulated | phenylacetaldehyde dehydrogenase | nitrate/nitrite transporter | nitrate reductase 2 (NRZ), gamma subunit | nitrate reductase 2 (NRZ), delta subunit (assembly subunit) | nitrate reductase 2 (NRZ), beta subunit | nitrate reductase 2 (NRZ), alpha subunit | serine/threonine-specific protein phosphatase 1 | ornithine decarboxylase isozyme, inducible | predicted inner membrane protein | conserved inner membrane protein | cardiolipin synthase 2 | predicted DNase | conserved protein | predicted protein | predicted enzyme | conserved protein with FAD/NAD(P)-binding domain | predicted multidrug efflux system | predicted chaperone | predicted diguanylate cyclase | predicted phosphodiesterase | fused DNA-binding transcriptional dual regulator/O6-methylguanine-DNA methyltransferase | L-asparagine transporter | DNA-binding transcriptional dual regulator | predicted mechanosensitive channel |
| 67 | 42 | 329 | -1.281 | 0.358 |  |  | 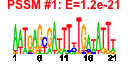 | 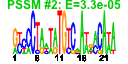 |  | b3721 b3723 b2269 b3898 b3558 b3593 b3482 b3659 b3119 b2858 b3043 b3120 b3121 b3483 b3490 b3520 b3562 b3563 b3596 b3595 b3765 b3818 b3861 b3889 b3896 b4038 b4047 b4048 b4145 b4204 b4325 b4347 b4552 b4205 b4215 b4017 b4455 b3557 b2850 b2856 b3615 b2849 | bglB bglG elaD frvX insK rhsA rhsB setC tdcR ygeN ygiL yhaB yhaC yhhH yhjB yiaA yiaB yibG yibJ yifB yigG yihF yiiE yiiG yjbI yjbL yjbM yjeJ yjfZ yjiC yjiW yrhC ytfA ytfI arpA hokA insJ ygeF ygeK yibD yqeK | cryptic phospho-beta-glucosidase B | transcriptional antiterminator of the bgl operon | predicted enzyme | predicted endo-1,4-beta-glucanase | IS150 conserved protein InsB | rhsA element core protein RshA | rhsB element core protein RshB | predicted sugar efflux system | DNA-binding transcriptional activator | predicted fimbrial-like adhesin protein | predicted protein | predicted DNA-binding response regulator in two-component regulatory system | conserved inner membrane protein | conserved protein | predicted bifunctional enzyme and transcriptional regulator | predicted inner membrane protein | predicted transcriptional regulator | ankyrin repeat protein, function unknown | toxic polypeptide, small | IS150 protein InsA | predicted glycosyl transferase |
| 48 | 36 | 331 | -1.265 | 0.348 |  |  | 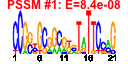 | 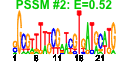 |  | b1731 b1737 b1734 b1733 b1735 b1579 b1463 b1845 b1581 b1580 b0913 b1063 b1325 b1432 b1462 b1542 b1694 b1695 b1697 b1699 b1771 b1772 b1774 b1775 b1776 b2002 b1368 b1436 b0982 b0944 b0943 b0981 b0983 b0984 b0985 b0986 | cedA chbC chbF chbG chbR intQ nhoA ptrB rspA rspB ycaI yceB ycjG ydcM yddH ydfI ydiF ydiO ydiQ ydiS ydjG ydjH ydjJ ydjK ydjL yeeS ynaA yncJ etp ycbF ycbV etk gfcE gfcD gfcC gfcB | cell division modulator | N,N'-diacetylchitobiose-specific enzyme IIC component of PTS | cryptic phospho-beta-glucosidase, NAD(P)-binding | conserved protein | DNA-binding transcriptional dual regulator | N-hydroxyarylamine O-acetyltransferase | protease II | predicted dehydratase | predicted oxidoreductase, Zn-dependent and NAD(P)-binding | conserved inner membrane protein | predicted lipoprotein | L-Ala-D/L-Glu epimerase | predicted transposase | predicted mannonate dehydrogenase | fused predicted acetyl-CoA:acetoacetyl-CoA transferase: alpha subunit/beta subunit | predicted acyl-CoA dehydrogenase | predicted oxidoreductase with FAD/NAD(P)-binding domain | predicted oxidoreductase | predicted kinase | predicted transporter | CP4-44 prophage; predicted DNA repair protein | predicted protein | phosphotyrosine-protein phosphatase | predicted periplasmic pilini chaperone | predicted fimbrial-like adhesin protein | cryptic autophosphorylating protein tyrosine kinase Etk | predicted exopolysaccharide export protein | predicted outer membrane lipoprotein |
| 143 | 50 | 337 | -1.254 | 0.579 |  |  |  | 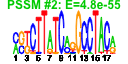 |  | b4016 b2219 b0038 b0037 b2451 b3081 b2485 b0698 b0697 b0696 b0695 b0694 b4513 b4037 b0353 b4180 b4431 b0997 b2045 b2043 b0223 b0228 b0323 b0324 b0445 b0689 b1050 b1110 b1471 b1741 b2128 b2129 b2130 b2131 b2245 b2354 b2389 b2681 b2812 b3103 b3662 b3698 b3901 b3944 b3954 b4030 b0258 b4509 b4534 b1973 | aceK atoS caiB caiC eutA fadH hyfE kdpA kdpB kdpC kdpD kdpE kdpF malM mhpT rlmB torA wcaK wcaM yafJ yafM yahI yahJ ybaE ybfP yceK ycfJ yddK cho yehW yehX yehY osmF yfaU yfdK yfeO ygaY ygdL yhaH nepI yidB rhaM yijF yijO psiE ykfC ylcG yodA | isocitrate dehydrogenase kinase/phosphatase | sensory histidine kinase in two-component regulatory system with AtoC | crotonobetainyl CoA:carnitine CoA transferase | predicted crotonobetaine CoA ligase:carnitine CoA ligase | reactivating factor for ethanolamine ammonia lyase | 2,4-dienoyl-CoA reductase, NADH and FMN-linked | hydrogenase 4, membrane subunit | potassium translocating ATPase, subunit A | potassium translocating ATPase, subunit B | potassium translocating ATPase, subunit C | fused sensory histidine kinase in two-component regulatory system with KdpE: signal sensing protein | DNA-binding response regulator in two-component regulatory system with KdpD | potassium ion accessory transporter subunit | maltose regulon periplasmic protein | predicted 3-hydroxyphenylpropionic transporter | 23S rRNA (Gm2251)-methyltransferase | trimethylamine N-oxide (TMAO) reductase I, catalytic subunit | predicted pyruvyl transferase | predicted colanic acid biosynthesis protein | predicted amidotransfease | conserved protein | predicted carbamate kinase-like protein | predicted deaminase with metallo-dependent hydrolase domain | predicted transporter subunit: periplasmic-binding component of ABC superfamily | predicted protein | predicted lipoprotein | endonuclease of nucleotide excision repair | predicted transporter subunit: membrane component of ABC superfamily | predicted transporter subunit: ATP-binding component of ABC superfamily | predicted 2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase | CPS-53 (KpLE1) prophage; conserved protein | predicted ion channel protein | predicted inner membrane protein | predicted transporter | L-rhamnose mutarotase | predicted DNA-binding transcriptional regulator | predicted phosphate starvation inducible protein | DLP12 prophage; predicted protein | conserved metal-binding protein |
| 19 | 48 | 334 | -1.197 | 0.409 |  |  | 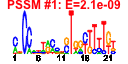 |  |  | b0263 b0262 b2715 b0615 b0107 b0347 b0548 b2051 b0108 b0236 b0550 b0561 b0246 b0248 b0251 b0317 b0318 b0319 b0320 b0321 b0547 b0549 b0771 b0823 b0824 b1010 b2164 b2165 b2357 b2358 b2634 b2643 b2644 b2769 b2770 b2771 b4463 b2774 b0249 b0247 b4504 b0245 b2139 b4548 b2915 b0825 b2356 b0250 | afuB afuC ascF citF hofB mhpA ninE gmm ppdD prfH rusA tfaD yafW yafX yafY yahC yahD yahE yahF yahG ybcN ybcO ybhJ ybiW ybiY rutC yeiM yeiN yfdN yfdO yfjR yfjX yfjY ygcQ ygcR ygcS ygcU ygcW ykfF ykfG ykfH ykfI mdtQ ypjJ yqfE fsaA yfdM ykfB | CP4-6 prophage; predicted ferric transporter subunit | fused cellobiose/arbutin/salicin-specific PTS enzymes: IIB component/IC component | citrate lyase, citrate-ACP transferase (alpha) subunit | conserved protein with nucleoside triphosphate hydrolase domain | 3-(3-hydroxyphenyl)propionate hydroxylase | DLP12 prophage; conserved protein | GDP-mannose mannosyl hydrolase | predicted major pilin subunit | DLP12 prophage; endonuclease RUS | CP4-6 prophage; antitoxin of the YkfI-YafW toxin-antitoxin system | CP4-6 prophage; predicted protein | CP4-6 prophage; predicted DNA-binding transcriptional regulator | predicted inner membrane protein | predicted transcriptional regulator with ankyrin domain | predicted protein | predicted acyl-CoA synthetase with NAD(P)-binding domain and succinyl-CoA synthetase domain | conserved protein | DLP12 prophage; predicted protein | predicted hydratase | predicted pyruvate formate lyase | predicted pyruvate formate lyase activating enzyme | predicted nucleoside transporter | CPS-53 (KpLE1) prophage; predicted protein | CP4-57 prophage; predicted DNA-binding transcriptional regulator | CP4-57 prophage; predicted antirestriction protein | CP4-57 prophage; predicted DNA repair protein | predicted flavoprotein | predicted transporter | predicted FAD containing dehydrogenase | predicted deoxygluconate dehydrogenase | CP4-6 prophage; predicted DNA repair protein | CP4-6 prophage; toxin of the YkfI-YafW toxin-antitoxin system | fructose-6-phosphate aldolase 1 |
| 86 | 48 | 332 | -1.113 | 0.399 |  |  |  |  |  | b2455 b2452 b2458 b2461 b2459 b1156 b2047 b0282 b0283 b0286 b1346 b2122 b2225 b2068 b0505 b2457 b2456 b2048 b2453 b2454 b2460 b0507 b0509 b0508 b2051 b2162 b0284 b0285 b0504 b2069 b2166 b2161 b2163 b2165 b2233 b2427 b2430 b2950 b1155 b4521 b2139 b2383 b2384 b2385 b2386 b2387 b2462 b2647 | eutE eutH eutD eutP eutT tfaE wcaJ yagP yagQ yagT ydaQ yehQ yfaP alkA allA eutM eutN cpsG eutG eutJ eutQ gcl glxR hyi gmm rihB yagR yagS allS yegD yeiC nupX yeiL yeiN yfaL yfeT yfeW yggR ymfS ycgI mdtQ fryA ypdE ypdF fryC fryB eutS ypjA | predicted aldehyde dehydrogenase, ethanolamine utilization protein | predicted inner membrane protein | predicted phosphotransacetylase subunit | conserved protein with nucleoside triphosphate hydrolase domain | predicted cobalamin adenosyltransferase in ethanolamine utilization | e14 prophage; predicted tail fiber assembly protein | predicted UDP-glucose lipid carrier transferase | predicted transcriptional regulator | conserved protein | predicted xanthine dehydrogenase, 2Fe-2S subunit | Rac prophage; conserved protein | 3-methyl-adenine DNA glycosylase II | ureidoglycolate hydrolase | predicted carboxysome structural protein, ethanolamine utilization protein | phosphomannomutase | predicted alcohol dehydrogenase in ethanolamine utilization | predicted chaperonin, ethanolamine utilization protein | glyoxylate carboligase | tartronate semialdehyde reductase, NADH-dependent | hydroxypyruvate isomerase | GDP-mannose mannosyl hydrolase | ribonucleoside hydrolase 2 | predicted oxidoreductase with molybdenum-binding domain | predicted oxidoreductase with FAD-binding domain | DNA-binding transcriptional activator of the allD operon | predicted chaperone | predicted kinase | predicted nucleoside transporter | DNA-binding transcriptional activator of stationary phase nitrogen survival | adhesin | predicted DNA-binding transcriptional regulator | predicted periplasmic esterase | predicted transporter | e14 prophage; predicted protein | fused predicted PTS enzymes: Hpr component/enzyme I component/enzyme IIA component | predicted peptidase | predicted enzyme IIC component of PTS | predicted enzyme IIB component of PTS | predicted carboxysome structural protein with predicted role in ethanol utilization | adhesin-like autotransporter |
| 7 | 64 | 328 | -1.103 | 0.421 | 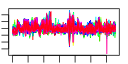 |  | 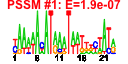 |  |  | b3665 b3140 b3801 b3722 b3720 b3693 b3691 b3900 b3899 b3897 b3683 b3681 b3323 b3324 b3325 b3326 b3327 b3328 b3329 b3330 b3331 b3332 b3333 b3334 b3335 b3379 b3322 b2825 b2823 b3904 b3906 b3905 b3907 b3566 b2824 b3078 b3104 b3269 b3376 b3377 b3378 b3380 b3381 b3442 b3518 b3575 b3576 b3577 b3578 b3656 b3657 b3664 b3678 b3679 b3680 b3684 b3718 b3944 b4181 b4182 b4184 b4185 b4257 b4485 | ade agaD aslA bglF bglH dgoK dgoT frvA frvB frvR glvC gspA gspC gspD gspE gspF gspG gspH gspI gspJ gspK gspL gspM gspO php gspB ppdB ppdC rhaB rhaR rhaS rhaT xylF ygdB ygjI yhaI yhdX yhfS yhfT yhfU yhfW yhfX yhhZ yhjA yiaK yiaL yiaM yiaN yicI yicJ yicO yidJ yidK yidL yidP yieK yijF yjfI yjfJ yjfL yjfM yjgN ytfR | cryptic adenine deaminase | N-acetylgalactosamine-specific enzyme IID component of PTS | acrylsulfatase-like enzyme | fused beta-glucoside-specific PTS enzymes: IIA component/IIB component/IIC component | carbohydrate-specific outer membrane porin, cryptic | 2-oxo-3-deoxygalactonate kinase | D-galactonate transporter | predicted enzyme IIA component of PTS | fused predicted PTS enzymes: IIB component/IIC component | predicted regulator | general secretory pathway component, cryptic | pseudopilin, cryptic, general secretion pathway | predicted general secretory pathway component, cryptic | bifunctional prepilin leader peptidase/ methylase | predicted hydrolase | part of gsp divergon involved in type II protein secretion | conserved protein | predicted protein | rhamnulokinase | DNA-binding transcriptional activator, L-rhamnose-binding | L-rhamnose:proton symporter | D-xylose transporter subunit | predicted transporter | predicted inner membrane protein | predicted amino-acid transporter subunit | predicted mutase | predicted amino acid racemase | predicted cytochrome C peroxidase | 2,3-diketo-L-gulonate dehydrogenase, NADH-dependent | predicted alpha-glucosidase | predicted xanthine/uracil permase | predicted sulfatase/phosphatase | predicted DNA-binding transcriptional regulator | predicted 6-phosphogluconolactonase | predicted transcriptional regulator effector protein | conserved inner membrane protein | predicted sugar transporter subunit: ATP-binding component of ABC superfamily |
| 114 | 56 | 331 | -1.052 | 0.391 | 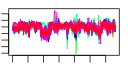 |  | 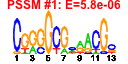 | 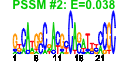 |  | b3932 b3811 b3343 b3345 b3346 b3731 b3732 b4052 b3613 b2927 b3729 b3730 b3424 b3413 b3612 b3779 b3608 b3610 b3805 b3804 b3803 b3802 b3931 b3187 b3350 b3351 b3998 b3784 b3791 b3787 b3786 b3788 b3780 b3782 b3832 b3649 b3609 b3349 b3650 b3836 b3839 b4483 b3651 b3781 b3835 b3792 b3793 b3785 b3344 b4551 b3611 b3614 b3812 b3834 b3859 b3999 | hslV xerC yheL yheN yheO atpC atpD dnaB envC epd glmS glmU glpG gntX gpmM gpp gpsA grxC hemC hemD hemX hemY hslU ispB kefB kefG nfi rfe rffA rffD rffE rffG rhlB rmuC rpoZ secB slyD spoT tatA tatC tatD trmH trxA ubiB wzxE wzyE wzzE yheM yheV yibN yibQ yigB yigP rdoA yjaG | peptidase component of the HslUV protease | site-specific tyrosine recombinase | predicted intracellular sulfur oxidation protein | predicted DNA-binding transcriptional regulator | F1 sector of membrane-bound ATP synthase, epsilon subunit | F1 sector of membrane-bound ATP synthase, beta subunit | replicative DNA helicase | protease with a role in cell division | D-erythrose 4-phosphate dehydrogenase | L-glutamine:D-fructose-6-phosphate aminotransferase | fused N-acetyl glucosamine-1-phosphate uridyltransferase/glucosamine-1-phosphate acetyl transferase | predicted intramembrane serine protease | gluconate periplasmic binding protein with phosphoribosyltransferase domain, GNT I system | phosphoglycero mutase III, cofactor-independent | guanosine pentaphosphatase/exopolyphosphatase | glycerol-3-phosphate dehydrogenase (NAD+) | glutaredoxin 3 | hydroxymethylbilane synthase | uroporphyrinogen III synthase | predicted uroporphyrinogen III methylase | predicted protoheme IX synthesis protein | molecular chaperone and ATPase component of HslUV protease | octaprenyl diphosphate synthase | potassium:proton antiporter | component of potassium effux complex with KefB | endonuclease V | UDP-GlcNAc:undecaprenylphosphate GlcNAc-1-phosphate transferase | TDP-4-oxo-6-deoxy-D-glucose transaminase | UDP-N-acetyl-D-mannosaminuronic acid dehydrogenase | UDP-N-acetyl glucosamine-2-epimerase | dTDP-glucose 4,6-dehydratase | ATP-dependent RNA helicase | predicted recombination limiting protein | RNA polymerase, omega subunit | protein export chaperone | FKBP-type peptidyl prolyl cis-trans isomerase (rotamase) | bifunctional (p)ppGpp synthetase II/ guanosine-3',5'-bis pyrophosphate 3'-pyrophosphohydrolase | TatABCE protein translocation system subunit | DNase, magnesium-dependent | tRNA (Guanosine-2'-O-)-methyltransferase | thioredoxin 1 | 2-octaprenylphenol hydroxylase | O-antigen translocase | predicted Wzy protein involved in ECA polysaccharide chain elongation | Entobacterial Common Antigen (ECA) polysaccharide chain length modulation protein | predicted intraceullular sulfur oxidation protein | predicted protein | predicted rhodanese-related sulfurtransferase | predicted polysaccharide deacetylase | predicted hydrolase | conserved protein | Thr/Ser kinase implicated in Cpx stress response |
| 127 | 42 | 328 | -0.834 | 0.391 | 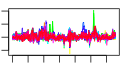 |  | 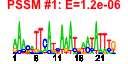 | 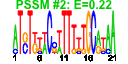 |  | b0555 b1789 b2359 b1788 b0587 b1352 b0568 b0569 b1351 b1349 b0700 b0933 b0365 b0366 b0367 b0368 b0289 b0290 b0291 b0292 b0567 b0644 b0645 b0681 b0682 b0719 b0709 b0716 b0717 b0718 b0938 b0939 b0940 b0941 b0942 b1239 b1318 b2070 b2361 b2362 b2363 b1527 | ybcS yeaL yfdP yoaI fepE kilR nfrA nfrB racC recT rhsC ssuB tauA tauB tauC tauD yagV yagW yagX matC ybcH ybeQ ybeR ybfM ybfN ybgD ybgH ybgO ybgP ybgQ ycbQ ycbR ycbS ycbT ycbU yegI yfdR yfdS yfdT yneK | DLP12 prophage; predicted lysozyme | conserved inner membrane protein | CPS-53 (KpLE1) prophage; predicted protein | predicted protein | regulator of length of O-antigen component of lipopolysaccharide chains | Rac prophage; inhibitor of ftsZ, killing protein | bacteriophage N4 receptor, outer membrane subunit | bacteriophage N4 receptor, inner membrane subunit | Rac prophage; predicted protein | Rac prophage; recombination and repair protein | rhsC element core protein RshC | alkanesulfonate transporter subunit | taurine transporter subunit | taurine dioxygenase, 2-oxoglutarate-dependent | conserved protein | predicted receptor | predicted aromatic compound dioxygenase | predicted outer membrane porin | predicted lipoprotein | predicted fimbrial-like adhesin protein | predicted transporter | predicted assembly protein | predicted outer membrane protein | predicted periplasmic pilin chaperone | predicted outer membrane usher protein | CPS-53 (KpLE1) prophage; conserved protein |
| 32 | 52 | 335 | -0.803 | 0.409 |  |  | 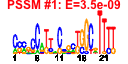 | 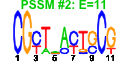 |  | b2457 b0518 b2540 b2826 b0292 b0324 b0513 b0551 b2298 b2355 b2430 b3882 b4026 b4509 b2383 b2384 b2385 b2386 b2387 b2466 b0476 b0263 b0262 b0514 b0107 b0106 b3069 b2429 b0548 b0108 b0550 b0066 b0246 b0248 b0317 b0318 b0319 b0320 b0321 b0547 b0549 b2164 b2634 b2643 b2644 b2872 b0253 b0249 b0247 b4504 b0245 b4548 | eutM fdrA hcaC ppdA matC yahJ ybbY ybcQ yfcC yfdL yfeW yihU yjbE ylcG fryA ypdE ypdF fryC fryB ypfG aes afuB afuC glxK hofB hofC ileX murP ninE ppdD rusA thiQ yafW yafX yahC yahD yahE yahF yahG ybcN ybcO yeiM yfjR yfjX yfjY ygeY ykfA ykfF ykfG ykfH ykfI ypjJ | predicted carboxysome structural protein, ethanolamine utilization protein | predicted acyl-CoA synthetase with NAD(P)-binding Rossmann-fold domain | 3-phenylpropionate dioxygenase, predicted ferredoxin subunit | conserved protein | predicted protein | predicted deaminase with metallo-dependent hydrolase domain | predicted uracil/xanthine transporter | DLP12 prophage; predicted antitermination protein | predicted inner membrane protein | predicted periplasmic esterase | predicted oxidoreductase with NAD(P)-binding Rossmann-fold domain | DLP12 prophage; predicted protein | fused predicted PTS enzymes: Hpr component/enzyme I component/enzyme IIA component | predicted peptidase | predicted enzyme IIC component of PTS | predicted enzyme IIB component of PTS | acetyl esterase | CP4-6 prophage; predicted ferric transporter subunit | glycerate kinase II | conserved protein with nucleoside triphosphate hydrolase domain | assembly protein in type IV pilin biogenesis, transmembrane protein | tRNA-Ile | fused predicted PTS enzymes: IIB component/IIC component | DLP12 prophage; conserved protein | predicted major pilin subunit | DLP12 prophage; endonuclease RUS | thiamin transporter subunit | CP4-6 prophage; antitoxin of the YkfI-YafW toxin-antitoxin system | CP4-6 prophage; predicted protein | predicted transcriptional regulator with ankyrin domain | predicted acyl-CoA synthetase with NAD(P)-binding domain and succinyl-CoA synthetase domain | predicted nucleoside transporter | CP4-57 prophage; predicted DNA-binding transcriptional regulator | CP4-57 prophage; predicted antirestriction protein | CP4-57 prophage; predicted DNA repair protein | CP4-6 prophage; predicted GTP-binding protein | CP4-6 prophage; predicted DNA repair protein | CP4-6 prophage; toxin of the YkfI-YafW toxin-antitoxin system |
| 82 | 57 | 330 | -0.778 | 0.400 |  |  | 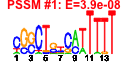 | 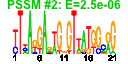 |  | b2918 b0507 b0508 b4102 b2920 b3878 b3880 b4028 b2775 b3139 b3141 b3136 b3501 b3374 b3950 b3949 b3953 b3137 b3952 b3951 b4105 b4101 b4100 b4099 b4098 b4097 b4096 b2879 b3061 b3062 b4193 b2881 b2238 b2769 b2770 b2771 b4463 b2774 b2919 b2878 b2880 b2886 b2887 b2983 b2986 b3063 b3080 b3211 b3589 b3879 b3881 b3883 b4027 b4080 b4081 b4082 b4358 | argK gcl hyi phnF scpC yihQ yihS yjbG yqcE agaC agaI agaS arsR frlD frwB frwC frwD kbaY pflC pflD phnD phnG phnH phnI phnJ phnK phnL ssnA ttdA ttdB ulaA xdhD yfaH ygcQ ygcR ygcS ygcU ygcW scpB ygfK ygfM ygfS ygfT yghQ yghT ttdT ygjK yhcC yiaY yihR yihT yihV yjbF mdtP mdtO mdtN yjjN | membrane ATPase/protein kinase | glyoxylate carboligase | hydroxypyruvate isomerase | predicted DNA-binding transcriptional regulator of phosphonate uptake and biodegradation | propionyl-CoA:succinate-CoA transferase | alpha-glucosidase | predicted glucosamine isomerase | conserved protein | predicted transporter | N-acetylgalactosamine-specific enzyme IIC component of PTS | galactosamine-6-phosphate isomerase | tagatose-6-phosphate ketose/aldose isomerase | DNA-binding transcriptional repressor | fructoselysine 6-kinase | predicted enzyme IIB component of PTS | predicted enzyme IIC component of PTS | tagatose 6-phosphate aldolase 1, kbaY subunit | pyruvate formate lyase II activase | predicted formate acetyltransferase 2 (pyruvate formate lyase II) | phosphonate/organophosphate ester transporter subunit | carbon-phosphorus lyase complex subunit | predicted chlorohydrolase/aminohydrolase | L-tartrate dehydratase, alpha subunit | L-tartrate dehydratase, beta subunit | L-ascorbate-specific enzyme IIC component of PTS | fused predicted xanthine/hypoxanthine oxidase: molybdopterin-binding subunit/Fe-S binding subunit | predicted flavoprotein | predicted FAD containing dehydrogenase | predicted deoxygluconate dehydrogenase | methylmalonyl-CoA decarboxylase, biotin-independent | predicted oxidoreductase, Fe-S subunit | predicted oxidoreductase | predicted oxidoreductase, 4Fe-4S ferredoxin-type subunit | fused predicted oxidoreductase: Fe-S subunit/nucleotide-binding subunit | predicted inner membrane protein | predicted protein with nucleoside triphosphate hydrolase domain | predicted tartrate:succinate antiporter | predicted glycosyl hydrolase | predicted Fe-S oxidoreductase | predicted Fe-containing alcohol dehydrogenase | predicted aldose-1-epimerase | predicted aldolase | predicted sugar kinase | predicted lipoprotein | predicted outer membrane factor of efflux pump | predicted multidrug efflux system component | predicted membrane fusion protein of efflux pump | predicted oxidoreductase, Zn-dependent and NAD(P)-binding |
| 120 | 60 | 321 | -0.755 | 0.381 |  |  | 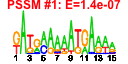 | 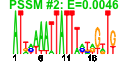 |  | b2485 b2490 b0998 b1168 b1318 b2003 b2005 b2070 b2121 b2226 b2229 b1409 b2222 b2224 b2221 b2223 b2368 b2367 b2455 b2452 b2458 b2461 b2459 b2481 b2483 b2486 b2487 b2488 b2491 b2873 b1348 b2711 b2519 b2987 b1347 b2071 b2108 b2109 b2110 b2118 b2120 b2122 b2123 b2230 b2225 b2270 b2271 b2272 b2333 b2334 b2335 b2336 b2339 b2636 b2637 b2870 b2871 b1506 b2547 b2548 | hyfE hyfJ torD ycgG yeeT yeeV yegI yehP yfaQ yfaT ynbB atoA atoB atoD atoE emrK emrY eutE eutH eutD eutP eutT hyfA hyfC hyfF hyfG hyfH hyfR hyuA lar norW pbpC pitB ydaC yegJ yehA yehB yehC yehI yehM yehQ yehR yfaA yfaP yfbK yfbL yfbM yfcP yfcQ yfcR yfcS yfcV yfjS yfjT ygeW ygeX yneL yphE yphF | hydrogenase 4, membrane subunit | predicted processing element hydrogenase 4 | chaperone involved in maturation of TorA subunit of trimethylamine N-oxide reductase system I | conserved inner membrane protein | CP4-44 prophage; predicted protein | CP4-44 prophage; toxin of the YeeV-YeeU toxin-antitoxin system | conserved protein | predicted protein | predicted CDP-diglyceride synthase | acetyl-CoA:acetoacetyl-CoA transferase, beta subunit | acetyl-CoA acetyltransferase | acetyl-CoA:acetoacetyl-CoA transferase, alpha subunit | short chain fatty acid transporter | EmrKY-TolC multidrug resistance efflux pump, membrane fusion protein component | predicted multidrug efflux system | predicted aldehyde dehydrogenase, ethanolamine utilization protein | predicted inner membrane protein | predicted phosphotransacetylase subunit | conserved protein with nucleoside triphosphate hydrolase domain | predicted cobalamin adenosyltransferase in ethanolamine utilization | hydrogenase 4, 4Fe-4S subunit | hydrogenase 4, subunit | hydrogenase 4, Fe-S subunit | DNA-binding transcriptional activator, formate sensing | D-stereospecific phenylhydantoinase | Rac prophage; restriction alleviation protein | NADH:flavorubredoxin oxidoreductase | fused transglycosylase/transpeptidase | phosphate transporter | Rac prophage; predicted protein | predicted fimbrial-like adhesin protein | predicted outer membrane protein | predicted periplasmic pilin chaperone | predicted peptidase | predicted periplasmic pilus chaperone | CP4-57 prophage; predicted protein | 2,3-diaminopropionate ammonia-lyase | fused predicted sugar transporter subunits of ABC superfamily: ATP-binding components | predicted sugar transporter subunit: periplasmic-binding component of ABC superfamily |
| 47 | 55 | 335 | -0.715 | 0.400 |  |  | 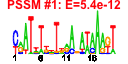 |  |  | b0517 b2624 b0040 b0297 b3076 b0041 b0042 b0043 b0044 b3374 b3952 b4105 b0331 b0333 b0334 b0335 b0330 b3581 b4193 b0045 b0252 b0290 b0291 b0363 b4462 b2735 b2736 b2737 b2738 b2739 b4464 b2886 b2887 b4465 b3079 b3080 b3220 b3874 b3879 b3881 b3883 b4296 b3144 b3145 b0036 b3370 b4106 b0561 b4196 b0013 b0011 b0251 b2654 b3880 b3882 | allD alpA caiT eaeH ebgA fixA fixB fixC fixX frlD pflC phnD prpB prpC prpD prpE prpR sgbH ulaA yaaU yafZ yagW yagX yaiP ygaQ ygbI ygbJ ygbK ygbL ygbM ygfQ ygfS ygfT yggP ygjJ ygjK yhcG yihN yihR yihT yihV yjhF yraJ yraK caiD frlA phnC tfaD ulaD yaaI yaaW yafY yihS yihU | ureidoglycolate dehydrogenase | CP4-57 prophage; DNA-binding transcriptional activator | predicted transporter | cryptic beta-D-galactosidase, alpha subunit | predicted electron transfer flavoprotein subunit, ETFP adenine nucleotide-binding domain | predicted electron transfer flavoprotein, NAD/FAD-binding domain and ETFP adenine nucleotide-binding domain-like protein | predicted oxidoreductase with FAD/NAD(P)-binding domain | predicted 4Fe-4S ferredoxin-type protein | fructoselysine 6-kinase | pyruvate formate lyase II activase | phosphonate/organophosphate ester transporter subunit | 2-methylisocitrate lyase | 2-methylcitrate synthase | 2-methylcitrate dehydratase | predicted propionyl-CoA synthetase with ATPase domain | DNA-binding transcriptional activator | 3-keto-L-gulonate 6-phosphate decarboxylase | L-ascorbate-specific enzyme IIC component of PTS | CP4-6 prophage; conserved protein | predicted receptor | predicted aromatic compound dioxygenase | predicted glucosyltransferase | predicted DNA-binding transcriptional regulator | predicted dehydrogenase, with NAD(P)-binding Rossmann-fold domain | conserved protein | predicted class II aldolase | predicted oxidoreductase, 4Fe-4S ferredoxin-type subunit | fused predicted oxidoreductase: Fe-S subunit/nucleotide-binding subunit | predicted dehydrogenase | predicted glycosyl hydrolase | predicted aldose-1-epimerase | predicted aldolase | predicted sugar kinase | KpLE2 phage-like element; predicted transporter | predicted outer membrane protein | predicted fimbrial-like adhesin protein | crotonobetainyl CoA hydratase | predicted fructoselysine transporter | predicted protein | CP4-6 prophage; predicted DNA-binding transcriptional regulator | predicted glucosamine isomerase | predicted oxidoreductase with NAD(P)-binding Rossmann-fold domain |
| 74 | 64 | 335 | -0.710 | 0.417 |  |  |  |  |  | b4067 b1002 b1415 b1883 b1884 b0598 b1189 b1190 b2365 b1879 b2151 b2241 b2243 b2149 b2148 b0764 b0679 b3225 b3224 b1421 b4512 b4068 b4069 b3072 b1888 b1887 b1882 b1881 b3076 b1880 b1891 b1892 b1878 b1923 b1937 b1940 b1941 b1944 b1947 b1949 b1950 b1926 b1566 b2242 b3426 b2239 b2240 b2150 b0763 b0765 b1890 b1889 b3222 b1951 b1885 b1886 b4355 b1742 b1904 b3525 b4110 b4109 b1044 b1760 | actP agp aldA cheB cheR cstA dadA dadX dsdX flhA galS glpA glpC mglA mglC modB nagE nanA nanT trg ybdD yjcH acs aer cheA cheW cheY cheZ ebgA flhB flhC flhD flhE fliC fliE fliH fliI fliL fliO fliQ fliR fliT flxA glpB glpD glpQ glpT mglB modA modC motA motB nanK rcsA tap tar tsr ves yecR yhjH yjcZ yjdA ymdA ynjH | acetate transporter | glucose-1-phosphatase/inositol phosphatase | aldehyde dehydrogenase A, NAD-linked | fused chemotaxis regulator: protein-glutamate methylesterase in two-component regulatory system with CheA | chemotaxis regulator, protein-glutamate methyltransferase | carbon starvation protein | D-amino acid dehydrogenase | alanine racemase 2, PLP-binding | predicted transporter | predicted flagellar export pore protein | DNA-binding transcriptional repressor | sn-glycerol-3-phosphate dehydrogenase (anaerobic), large subunit, FAD/NAD(P)-binding | sn-glycerol-3-phosphate dehydrogenase (anaerobic), small subunit | fused methyl-galactoside transporter subunits of ABC superfamily: ATP-binding components | methyl-galactoside transporter subunit | molybdate transporter subunit | fused N-acetyl glucosamine specific PTS enzyme: IIC, IIB , and IIA components | N-acetylneuraminate lyase | sialic acid transporter | methyl-accepting chemotaxis protein III, ribose and galactose sensor receptor | conserved protein | conserved inner membrane protein involved in acetate transport | acetyl-CoA synthetase | fused signal transducer for aerotaxis sensory component/methyl accepting chemotaxis component | fused chemotactic sensory histidine kinase in two-component regulatory system with CheB and CheY: sensory histidine kinase/signal sensing protein | purine-binding chemotaxis protein | chemotaxis regulator transmitting signal to flagellar motor component | chemotaxis regulator, protein phosphatase for CheY | cryptic beta-D-galactosidase, alpha subunit | DNA-binding transcriptional dual regulator with FlhD | DNA-binding transcriptional dual regulator with FlhC | flagellar filament structural protein (flagellin) | flagellar basal-body component | flagellar biosynthesis protein | flagellum-specific ATP synthase | flagellar export pore protein | predicted chaperone | Qin prophage; predicted protein | sn-glycerol-3-phosphate dehydrogenase (anaerobic), membrane anchor subunit | sn-glycerol-3-phosphate dehydrogenase, aerobic, FAD/NAD(P)-binding | periplasmic glycerophosphodiester phosphodiesterase | sn-glycerol-3-phosphate transporter | proton conductor component of flagella motor | protein that enables flagellar motor rotation | predicted N-acetylmannosamine kinase | DNA-binding transcriptional activator, co-regulator with RcsB | methyl-accepting protein IV | methyl-accepting chemotaxis protein II | methyl-accepting chemotaxis protein I, serine sensor receptor | predicted protein | EAL domain containing protein involved in flagellar function | conserved protein with nucleoside triphosphate hydrolase domain |
| 123 | 59 | 337 | -0.697 | 0.437 |  |  | 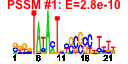 | 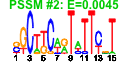 |  | b3871 b0470 b2569 b2568 b0182 b3258 b2567 b0183 b2576 b2533 b2559 b2952 b2954 b0185 b0910 b0184 b0173 b0420 b4410 b0180 b0028 b0172 b0175 b3425 b3560 b3559 b3181 b2514 b0178 b0125 b0026 b0421 b2515 b0174 b0181 b0179 b0027 b3018 b3164 b0025 b2570 b0098 b0408 b3981 b0409 b3175 b0423 b0949 b0422 b0176 b0177 b0948 b2516 b2512 b2513 b2532 b4469 b4049 b2689 | typA dnaX lepA lepB lpxB panF rnc rnhB srmB suhB tadA yggT rdgB accA cmk dnaE dxr dxs ecnA fabZ fkpB frr cdsA glpE glyQ glyS greA hisS skp hpt ileS ispA ispG ispU lpxA lpxD lspA plsC pnp ribF rseC secA secD secE secF secG thiI uup xseB rseP yaeT rlmL yfgA yfgL yfgM trmJ ygiQ dusA yqaA | GTP-binding protein | DNA polymerase III/DNA elongation factor III, tau and gamma subunits | GTP-binding membrane protein | leader peptidase (signal peptidase I) | tetraacyldisaccharide-1-P synthase | pantothenate:sodium symporter | RNase III | ribonuclease HII, degrades RNA of DNA-RNA hybrids | ATP-dependent RNA helicase | inositol monophosphatase | tRNA-specific adenosine deaminase | predicted inner membrane protein | dITP/XTP pyrophosphatase | acetyl-CoA carboxylase, carboxytransferase, alpha subunit | cytidylate kinase | DNA polymerase III alpha subunit | 1-deoxy-D-xylulose 5-phosphate reductoisomerase | 1-deoxyxylulose-5-phosphate synthase, thiamine-requiring, FAD-requiring | entericidin A membrane lipoprotein, antidote entericidin B | (3R)-hydroxymyristol acyl carrier protein dehydratase | FKBP-type peptidyl-prolyl cis-trans isomerase (rotamase) | ribosome recycling factor | CDP-diglyceride synthase | thiosulfate:cyanide sulfurtransferase (rhodanese) | glycine tRNA synthetase, alpha subunit | glycine tRNA synthetase, beta subunit | transcription elongation factor | histidyl tRNA synthetase | periplasmic chaperone | hypoxanthine phosphoribosyltransferase | isoleucyl-tRNA synthetase | geranyltranstransferase | 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase | undecaprenyl pyrophosphate synthase | UDP-N-acetylglucosamine acetyltransferase | UDP-3-O-(3-hydroxymyristoyl)-glucosamine N-acyltransferase | prolipoprotein signal peptidase (signal peptidase II) | 1-acyl-sn-glycerol-3-phosphate acyltransferase | polynucleotide phosphorylase/polyadenylase | bifunctional riboflavin kinase/FAD synthetase | RseC protein involved in reduction of the SoxR iron-sulfur cluster | preprotein translocase subunit, ATPase | SecYEG protein translocase auxillary subunit | preprotein translocase membrane subunit | sulfurtransferase required for thiamine and 4-thiouridine biosynthesis | fused predicted transporter subunits of ABC superfamily: ATP-binding components | exonuclease VII small subunit | zinc metallopeptidase | conserved protein | predicted methyltransferase | protein assembly complex, lipoprotein component | tRNA-dihydrouridine synthase A | conserved inner membrane protein |
| 54 | 58 | 331 | -0.618 | 0.423 |  |  |  |  |  | b2893 b2895 b3741 b3740 b4259 b4348 b3618 b3687 b3686 b4346 b3742 b1109 b3397 b4260 b4108 b3493 b2679 b3842 b3623 b4425 b3348 b3965 b4258 b2894 b0232 b0413 b2605 b2604 b3470 b3492 b3859 b4261 b4262 b2939 b3033 b3743 b3347 b3844 b3360 b2677 b2678 b3628 b3621 b3631 b3627 b3626 b3630 b3632 b3629 b3625 b3624 b0414 b2938 b3843 b3705 b4557 b3755 b3042 | dsbC fldB mnmG gidB holC hsdS htrL ibpA ibpB mcrB mioC ndh nudE pepA yjdM pitA proX rfaH waaU slyX trmA valS xerD yafN nrdR yfiB yfiN sirA yhiN rdoA yjgP yjgQ yqgB yqiB asnC fkpA fre pabA proV proW rfaB rfaC rfaG rfaI rfaJ rfaP rfaQ rfaS rfaY rfaZ ribD speA ubiD yidC yidD yieP yqiC | protein disulfide isomerase II | flavodoxin 2 | glucose-inhibited cell-division protein | methyltransferase, SAM-dependent methyltransferase, glucose-inhibited cell-division protein | DNA polymerase III, chi subunit | specificity determinant for hsdM and hsdR | predicted protein | heat shock chaperone | 5-methylcytosine-specific restriction enzyme McrBC, subunit McrB | FMN-binding protein MioC | respiratory NADH dehydrogenase 2/cupric reductase | ADP-ribose diphosphatase | aminopeptidase A, a cyteinylglycinase | conserved protein | phosphate transporter, low-affinity | glycine betaine transporter subunit | DNA-binding transcriptional antiterminator | lipopolysaccharide core biosynthesis | tRNA (uracil-5-)-methyltransferase | valyl-tRNA synthetase | site-specific tyrosine recombinase | predicted antitoxin of the YafO-YafN toxin-antitoxin system | predicted outer membrane lipoprotein | predicted diguanylate cyclase | conserved protein required for cell growth | predicted oxidoreductase with FAD/NAD(P)-binding domain | Thr/Ser kinase implicated in Cpx stress response | conserved inner membrane protein | predicted dehydrogenase | DNA-binding transcriptional dual regulator | FKBP-type peptidyl-prolyl cis-trans isomerase (rotamase) | flavin reductase | aminodeoxychorismate synthase, subunit II | UDP-D-galactose:(glucosyl)lipopolysaccharide-1, 6-D-galactosyltransferase | ADP-heptose:LPS heptosyl transferase I | glucosyltransferase I | UDP-D-galactose:(glucosyl)lipopolysaccharide- alpha-1,3-D-galactosyltransferase | UDP-D-glucose:(galactosyl)lipopolysaccharide glucosyltransferase | kinase that phosphorylates core heptose of lipopolysaccharide | lipopolysaccharide core biosynthesis protein | fused diaminohydroxyphosphoribosylaminopyrimidine deaminase and 5-amino-6-(5-phosphoribosylamino) uracil reductase | biosynthetic arginine decarboxylase, PLP-binding | 3-octaprenyl-4-hydroxybenzoate decarboxylase | cytoplasmic insertase into membrane protein, Sec system | predicted transcriptional regulator |
| 69 | 65 | 332 | -0.569 | 0.426 |  |  | 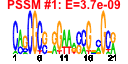 | 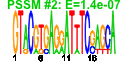 |  | b3256 b2316 b1866 b0420 b2566 b1092 b1091 b0684 b0683 b2036 b1132 b1712 b0421 b0642 b1116 b0222 b0181 b0924 b1865 b0416 b1714 b1090 b1831 b2565 b2039 b2038 b2040 b2037 b0415 b1084 b1863 b0408 b0687 b0921 b0423 b1719 b0737 b0738 b0949 b2035 b2034 b2033 b2032 b0422 b0407 b0685 b0948 b1864 b2350 b2351 b2513 b2554 b2555 b2611 b2948 b2949 b0661 b0923 b0922 b1713 b2585 b2041 b0947 b2187 b2188 | accC accD aspS dxs era fabD fabH fldA fur glf hflD ihfA ispA leuS lolC lpcA lpxA mukB nudB nusB pheS plsX proQ recO rfbA rfbC rfbD rfbX ribE rne ruvC secD seqA smtA thiI thrS tolQ tolR uup rfc wbbI wbbJ wbbK xseB yajC ybfE rlmL yebC yfdG yfdH yfgM yfhA yfhG ypjD yqgE yqgF miaB mukE mukF pheT pssA rfbB ycbX yejL yejM | acetyl-CoA carboxylase, biotin carboxylase subunit | acetyl-CoA carboxylase, beta (carboxyltranferase) subunit | aspartyl-tRNA synthetase | 1-deoxyxylulose-5-phosphate synthase, thiamine-requiring, FAD-requiring | membrane-associated, 16S rRNA-binding GTPase | malonyl-CoA-[acyl-carrier-protein] transacylase | 3-oxoacyl-[acyl-carrier-protein] synthase III | flavodoxin 1 | DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport | UDP-galactopyranose mutase, FAD/NAD(P)-binding | predicted lysogenization regulator | integration host factor (IHF), DNA-binding protein, alpha subunit | geranyltranstransferase | leucyl-tRNA synthetase | outer membrane-specific lipoprotein transporter subunit | D-sedoheptulose 7-phosphate isomerase | UDP-N-acetylglucosamine acetyltransferase | fused chromosome partitioning protein: predicted nucleotide hydrolase/conserved protein/conserved protein | dATP pyrophosphohydrolase | transcription antitermination protein | phenylalanine tRNA synthetase, alpha subunit | fatty acid/phospholipid synthesis protein | predicted structural transport element | gap repair protein | glucose-1-phosphate thymidylyltransferase | dTDP-4-deoxyrhamnose-3,5-epimerase | dTDP-4-dehydrorhamnose reductase subunit, NAD(P)-binding, of dTDP-L-rhamnose synthase | predicted polisoprenol-linked O-antigen transporter | riboflavin synthase beta chain | fused ribonucleaseE: endoribonuclease/RNA-binding protein/RNA degradosome binding protein | component of RuvABC resolvasome, endonuclease | SecYEG protein translocase auxillary subunit | regulatory protein for replication initiation | predicted S-adenosyl-L-methionine-dependent methyltransferase | sulfurtransferase required for thiamine and 4-thiouridine biosynthesis | threonyl-tRNA synthetase | membrane spanning protein in TolA-TolQ-TolR complex | fused predicted transporter subunits of ABC superfamily: ATP-binding components | O-antigen polymerase | conserved protein | predicted acyl transferase | lipopolysaccharide biosynthesis protein | exonuclease VII small subunit | lexA-regulated predicted protein | predicted methyltransferase | CPS-53 (KpLE1) prophage; bactoprenol-linked glucose translocase (flippase) | CPS-53 (KpLE1) prophage; bactoprenol glucosyl transferase | predicted DNA-binding response regulator in two-component system | predicted inner membrane protein | predicted protein | predicted Holliday junction resolvase | isopentenyl-adenosine A37 tRNA methylthiolase | protein involved in chromosome partitioning | Involved in chromosome partioning, Ca2+ binding protein | phenylalanine tRNA synthetase, beta subunit | phosphatidylserine synthase (CDP-diacylglycerol-serine O-phosphatidyltransferase) | dTDP-glucose 4,6 dehydratase, NAD(P)-binding | predicted 2Fe-2S cluster-containing protein | predicted hydrolase, inner membrane |
| 130 | 54 | 333 | -0.543 | 0.398 |  |  | 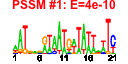 | 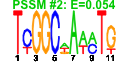 |  | b0617 b0616 b0613 b0614 b0353 b0692 b2004 b2470 b2252 b0512 b2257 b0618 b0615 b0612 b1043 b2482 b2490 b0731 b0732 b1377 b2407 b2406 b0117 b0511 b0527 b0702 b0704 b0703 b0768 b0769 b0770 b1115 b1168 b1196 b1311 b1576 b1689 b1691 b1801 b2226 b2229 b2253 b2254 b2255 b2256 b2258 b4544 b2359 b2408 b0563 b1455 b1974 b2105 b2106 | citD citE citG citX mhpT potE yeeU acrD ais allB arnT citC citF citT csgC hyfB hyfJ mngA mngB ompN xapA xapB yacH ybbW ybcI ybfB ybfC ybfO ybhD ybhH ybhI ycfT ycgG ycgY ycjO ydfD ydiL ydiN yeaV yfaQ yfaT arnB arnC arnA yfbH yfbJ yfbW yfdP yfeN tfaX yncH yodB rcnR rcnA | citrate lyase, acyl carrier (gamma) subunit | citrate lyase, citryl-ACP lyase (beta) subunit | triphosphoribosyl-dephospho-CoA transferase | apo-citrate lyase phosphoribosyl-dephospho-CoA transferase | predicted 3-hydroxyphenylpropionic transporter | putrescine/proton symporter: putrescine/ornithine antiporter | CP4-44 prophage; antitoxin of the YeeV-YeeU toxin-antitoxin system | aminoglycoside/multidrug efflux system | conserved protein | allantoinase | 4-amino-4-deoxy-L-arabinose transferase | citrate lyase synthetase | citrate lyase, citrate-ACP transferase (alpha) subunit | citrate:succinate antiporter | predicted curli production protein | hydrogenase 4, membrane subunit | predicted processing element hydrogenase 4 | fused 2-O-a-mannosyl-D-glycerate specific PTS enzymes: IIA component/IIB component/IIC component | alpha-mannosidase | outer membrane pore protein N, non-specific | purine nucleoside phosphorylase II | xanthosine transporter | predicted protein | predicted allantoin transporter | conserved inner membrane protein | predicted inner membrane protein | predicted DNA-binding transcriptional regulator | predicted transporter | predicted sugar transporter subunit: membrane component of ABC superfamily | Qin prophage; predicted protein | uridine 5'-(beta-1-threo-pentapyranosyl-4-ulose diphosphate) aminotransferase, PLP-dependent | undecaprenyl phosphate-L-Ara4FN transferase | fused UDP-L-Ara4N formyltransferase/UDP-GlcA C-4'-decarboxylase | CPS-53 (KpLE1) prophage; predicted protein | conserved outer membrane protein | predicted cytochrome | membrane protein conferring nickel and cobalt resistance |
| 111 | 60 | 332 | -0.531 | 0.406 |  |  | 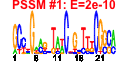 | 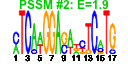 |  | b1767 b1418 b0954 b1651 b1210 b1215 b1048 b1049 b1650 b1186 b0688 b1768 b1204 b1281 b1286 b1584 b1238 b1763 b1637 b2066 b0686 b1035 b1203 b1233 b1213 b1253 b1254 b1255 b1765 b1780 b1839 b1875 b1869 b1870 b1295 b1583 b1727 b1809 b2027 b0887 b1288 b1236 b1842 b1829 b1208 b0918 b1209 b1638 b1129 b1830 b1211 b1212 b1207 b1232 b1764 b1034 b1214 b1282 b1928 b1726 | ansA cybB fabA gloA hemA kdsA mdoG mdoH nemA nhaB pgm pncA pth pyrF rnb speG tdk topB tyrS udk ybfF ycdY ychF ychJ ychQ yciA yciB yciC ydjA yeaD yebY yecM yecN cmoA ymjA ynfB yniC yoaB cld cydD fabI galU holE htpX ispE kdsB lolB pdxH phoQ prc prfA prmC prs purU selD ycdX ychA yciH yedD yniB | cytoplasmic L-asparaginase I | cytochrome b561 | beta-hydroxydecanoyl thioester dehydrase | glyoxalase I, Ni-dependent | glutamyl tRNA reductase | 3-deoxy-D-manno-octulosonate 8-phosphate synthase | glucan biosynthesis protein, periplasmic | glucan biosynthesis: glycosyl transferase | N-ethylmaleimide reductase, FMN-linked | sodium:proton antiporter | phosphoglucomutase | nicotinamidase/pyrazinamidase | peptidyl-tRNA hydrolase | orotidine-5'-phosphate decarboxylase | ribonuclease II | spermidine N1-acetyltransferase | thymidine kinase/deoxyuridine kinase | DNA topoisomerase III | tyrosyl-tRNA synthetase | uridine/cytidine kinase | conserved protein | predicted GTP-binding protein | predicted transcriptional regulator | predicted hydrolase | predicted inner membrane protein | predicted oxidoreductase | predicted protein | predicted metal-binding enzyme | predicted methyltransferase | regulator of length of O-antigen component of lipopolysaccharide chains | fused cysteine transporter subunits of ABC superfamily: membrane component/ATP-binding component | enoyl-[acyl-carrier-protein] reductase, NADH-dependent | glucose-1-phosphate uridylyltransferase | DNA polymerase III, theta subunit | predicted endopeptidase | 4-diphosphocytidyl-2-C-methylerythritol kinase | 3-deoxy-manno-octulosonate cytidylyltransferase | chaperone for lipoproteins | pyridoxine 5'-phosphate oxidase | sensory histidine kinase in two-compoent regulatory system with PhoP | carboxy-terminal protease for penicillin-binding protein 3 | peptide chain release factor RF-1 | N5-glutamine methyltransferase, modifies release factors RF-1 and RF-2 | phosphoribosylpyrophosphate synthase | formyltetrahydrofolate hydrolase | selenophosphate synthase | predicted zinc-binding hydrolase |
| 77 | 58 | 327 | -0.487 | 0.387 |  |  |  |  |  | b0474 b1093 b2400 b2508 b0884 b1208 b0918 b0630 b2890 b2114 b2113 b0914 b2320 b1830 b1211 b1212 b1207 b1277 b0893 b1133 b2318 b0888 b2319 b0853 b1214 b2171 b2595 b2316 b0632 b1092 b1091 b2315 b2042 b2036 b0680 b2029 b2507 b1132 b1116 b1782 b0924 b1090 b1831 b2039 b2038 b2040 b2037 b2498 b2035 b2034 b2033 b2032 b0407 b1832 b1840 b1983 b2351 b1841 | adk fabG gltX guaB infA ispE kdsB lipB lysS metG mrp msbA pdxB prc prfA prmC prs ribA serS mnmA truA trxB usg ybjN ychA yeiP yfiO accD dacA fabD fabH folC galF glf glnS gnd guaA hflD lolC mipA mukB plsX proQ rfbA rfbC rfbD rfbX upp rfc wbbI wbbJ wbbK yajC yebR yebZ yeeN yfdH yobA | adenylate kinase | 3-oxoacyl-[acyl-carrier-protein] reductase | glutamyl-tRNA synthetase | IMP dehydrogenase | translation initiation factor IF-1 | 4-diphosphocytidyl-2-C-methylerythritol kinase | 3-deoxy-manno-octulosonate cytidylyltransferase | lipoyl-protein ligase | lysine tRNA synthetase, constitutive | methionyl-tRNA synthetase | antiporter inner membrane protein | fused lipid transporter subunits of ABC superfamily: membrane component/ATP-binding component | erythronate-4-phosphate dehydrogenase | carboxy-terminal protease for penicillin-binding protein 3 | peptide chain release factor RF-1 | N5-glutamine methyltransferase, modifies release factors RF-1 and RF-2 | phosphoribosylpyrophosphate synthase | GTP cyclohydrolase II | seryl-tRNA synthetase, also charges selenocysteinyl-tRNA with serine | tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase | pseudouridylate synthase I | thioredoxin reductase, FAD/NAD(P)-binding | predicted semialdehyde dehydrogenase | predicted oxidoreductase | predicted transcriptional regulator | predicted elongtion factor | predicted lipoprotein | acetyl-CoA carboxylase, beta (carboxyltranferase) subunit | D-alanyl-D-alanine carboxypeptidase (penicillin-binding protein 5) | malonyl-CoA-[acyl-carrier-protein] transacylase | 3-oxoacyl-[acyl-carrier-protein] synthase III | bifunctional folylpolyglutamate synthase/ dihydrofolate synthase | predicted subunit with GalU | UDP-galactopyranose mutase, FAD/NAD(P)-binding | gluconate-6-phosphate dehydrogenase, decarboxylating | GMP synthetase (glutamine aminotransferase) | predicted lysogenization regulator | outer membrane-specific lipoprotein transporter subunit | scaffolding protein for murein synthesizing machinery | fused chromosome partitioning protein: predicted nucleotide hydrolase/conserved protein/conserved protein | fatty acid/phospholipid synthesis protein | predicted structural transport element | glucose-1-phosphate thymidylyltransferase | dTDP-4-deoxyrhamnose-3,5-epimerase | dTDP-4-dehydrorhamnose reductase subunit, NAD(P)-binding, of dTDP-L-rhamnose synthase | predicted polisoprenol-linked O-antigen transporter | uracil phosphoribosyltransferase | O-antigen polymerase | conserved protein | predicted acyl transferase | lipopolysaccharide biosynthesis protein | SecYEG protein translocase auxillary subunit | predicted inner membrane protein | CPS-53 (KpLE1) prophage; bactoprenol glucosyl transferase |
| 66 | 51 | 343 | -0.481 | 0.487 |  |  |  | 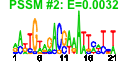 |  | b0715 b1475 b1476 b0348 b0349 b0350 b0352 b0351 b0346 b1814 b1743 b0289 b2292 b2671 b0308 b1171 b2390 b0774 b0777 b0778 b0776 b0034 b2731 b2000 b2552 b2725 b2724 b2723 b2722 b2721 b2720 b2719 b2718 b2717 b2489 b2726 b2729 b2730 b1936 b3942 b0347 b3756 b2591 b0201 b0293 b0919 b1971 b1972 b0836 b1172 b1757 | abrB fdnH fdnI mhpB mhpC mhpD mhpE mhpF mhpR sdaA spy yagV yfbS ygaC ykgG ymgD ypeC bioA bioC bioD bioF caiF fhlA flu hmp hycA hycB hycC hycD hycE hycF hycG hycH hycI hyfI hypA hypD hypE intG katG mhpA rrsC rrsG rrsH matB ycbJ yedY yedZ bssR ymgG ynjE | predicted regulator | formate dehydrogenase-N, Fe-S (beta) subunit, nitrate-inducible | formate dehydrogenase-N, cytochrome B556 (gamma) subunit, nitrate-inducible | 2,3-dihydroxyphenylpropionate 1,2-dioxygenase | 2-hydroxy-6-ketonona-2,4-dienedioic acid hydrolase | 2-keto-4-pentenoate hydratase | 4-hyroxy-2-oxovalerate/4-hydroxy-2-oxopentanoic acid aldolase, class I | acetaldehyde-CoA dehydrogenase II, NAD-binding | DNA-binding transcriptional activator, 3HPP-binding | L-serine deaminase I | envelope stress induced periplasmic protein | conserved protein | predicted transporter | predicted protein | 7,8-diaminopelargonic acid synthase, PLP-dependent | predicted methltransferase, enzyme of biotin synthesis | dethiobiotin synthetase | 8-amino-7-oxononanoate synthase | DNA-binding transcriptional activator | CP4-44 prophage; antigen 43 (Ag43) phase-variable biofilm formation autotransporter | fused nitric oxide dioxygenase/dihydropteridine reductase 2 | regulator of the transcriptional regulator FhlA | hydrogenase 3, Fe-S subunit | hydrogenase 3, membrane subunit | hydrogenase 3, large subunit | formate hydrogenlyase complex iron-sulfur protein | hydrogenase 3 and formate hydrogenase complex, HycG subunit | protein required for maturation of hydrogenase 3 | protease involved in processing C-terminal end of HycE | hydrogenase 4, Fe-S subunit | protein involved in nickel insertion into hydrogenases 3 | protein required for maturation of hydrogenases | carbamoyl phosphate phosphatase, hydrogenase 3 maturation protein | catalase/hydroperoxidase HPI(I) | 3-(3-hydroxyphenyl)propionate hydroxylase | 16S ribosomal RNA of rrnC operon | 16S ribosomal RNA of rrnG operon | 16S ribosomal RNA of rrnH operon | predicted reductase | conserved inner membrane protein | predicted thiosulfate sulfur transferase |
| 113 | 66 | 327 | -0.479 | 0.408 |  |  |  | 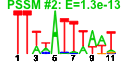 |  | b2071 b2339 b3721 b3723 b2269 b0535 b3875 b2854 b2648 b2734 b3906 b3659 b3119 b1156 b0135 b0278 b0279 b0326 b0364 b0558 b1001 b1309 b2273 b2274 b2275 b2345 b2375 b2625 b2626 b2645 b4462 b2851 b2852 b2853 b2855 b3120 b3121 b3173 b3215 b3216 b3504 b3562 b3563 b3818 b3861 b3896 b4038 b4047 b4048 b4066 b4204 b4325 b4365 b0310 b0294 b0370 b2376 b2650 b3013 b3046 b3048 b3142 b3143 b3144 b4205 b4215 | yegJ yfcV bglB bglG elaD fimZ ompL pbl pinH pphB rhaR setC tdcR tfaE yadC yagL yagM yahL yaiS ybcV yccE ycjM yfbN yfbO yfbP yfdF yfdX yfjI yfjJ yfjZ ygaQ ygeG ygeH ygeI yhaB yhaC yhbX yhcA yhcD yhiS yiaA yiaB yigG yihF yiiG yjbI yjbL yjbM yjcF yjfZ yjiC yjjQ ykgH matA ypdI ypjC yqhG yqiG yqiI yraH yraI yraJ ytfA ytfI | predicted protein | predicted fimbrial-like adhesin protein | cryptic phospho-beta-glucosidase B | transcriptional antiterminator of the bgl operon | predicted enzyme | predicted DNA-binding transcriptional regulator | predicted outer membrane porin L | serine/threonine-specific protein phosphatase 2 | DNA-binding transcriptional activator, L-rhamnose-binding | predicted sugar efflux system | DNA-binding transcriptional activator | e14 prophage; predicted tail fiber assembly protein | CP4-6 prophage; DNA-binding protein | CP4-6 prophage; predicted protein | conserved protein | DLP12 prophage; predicted protein | predicted glucosyltransferase | CP4-57 prophage; predicted protein | CP4-57 prophage; antitoxin of the YpjF-YfjZ toxin-antitoxin system | predicted chaperone | predictedtranscriptional regulator | predicted hydrolase, inner membrane | predicted periplasmic chaperone protein | predicted outer membrane protein | conserved inner membrane protein | predicted inner membrane protein | predicted regulator | predicted lipoprotein involved in colanic acid biosynthesis | predicted periplasmic pilin chaperone |
| 90 | 59 | 328 | -0.448 | 0.437 |  |  | 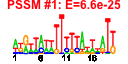 | 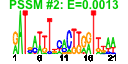 |  | b2252 b0875 b2257 b0612 b0229 b0731 b0732 b0532 b2407 b0567 b0527 b0644 b0645 b0691 b0682 b0703 b0768 b0769 b0770 b0938 b0939 b0940 b0942 b1196 b1691 b1730 b2253 b2254 b2255 b2256 b2258 b2506 b0563 b1028 b1042 b1041 b0338 b0339 b0341 b0646 b0649 b0566 b0730 b0530 b0531 b0534 b0533 b0227 b0385 b0544 b0545 b0551 b0562 b0601 b0602 b0647 b0648 b0876 b1029 | ais aqpZ arnT citT lfhA mngA mngB sfmD xapA ybcH ybcI ybeQ ybeR ybfG ybfN ybfO ybhD ybhH ybhI ycbQ ycbR ycbS ycbU ycgY ydiN ydjO arnB arnC arnA yfbH yfbJ yfgI tfaX ymdE csgA csgB cynR cynT cynX djlB djlC envY mngR sfmA sfmC sfmF sfmH yafL adrA ybcK ybcL ybcQ ybcY ybdM ybdN ybeT ybeU ybjD ycdU | conserved protein | aquaporin | 4-amino-4-deoxy-L-arabinose transferase | citrate:succinate antiporter | fused 2-O-a-mannosyl-D-glycerate specific PTS enzymes: IIA component/IIB component/IIC component | alpha-mannosidase | predicted outer membrane export usher protein | purine nucleoside phosphorylase II | predicted protein | conserved inner membrane protein | predicted lipoprotein | predicted DNA-binding transcriptional regulator | predicted transporter | predicted fimbrial-like adhesin protein | predicted periplasmic pilin chaperone | predicted outer membrane usher protein | uridine 5'-(beta-1-threo-pentapyranosyl-4-ulose diphosphate) aminotransferase, PLP-dependent | undecaprenyl phosphate-L-Ara4FN transferase | fused UDP-L-Ara4N formyltransferase/UDP-GlcA C-4'-decarboxylase | predicted inner membrane protein | cryptic curlin major subunit | curlin nucleator protein, minor subunit in curli complex | DNA-binding transcriptional dual regulator | carbonic anhydrase | predicted cyanate transporter | predicted chaperone | Hsc56 co-chaperone of HscC | DNA-binding transcriptional activator of porin biosynthesis | DNA-binding transcriptional dual regulator, fatty-acyl-binding | pilin chaperone, periplasmic | predicted lipoprotein and C40 family peptidase | predicted diguanylate cyclase | DLP12 prophage; predicted recombinase | DLP12 prophage; predicted kinase inhibitor | DLP12 prophage; predicted antitermination protein | conserved outer membrane protein | predicted tRNA ligase | conserved protein with nucleoside triphosphate hydrolase domain |
| 73 | 61 | 340 | -0.432 | 0.445 |  |  | 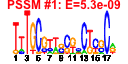 | 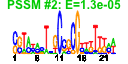 |  | b3475 b3973 b2143 b3911 b3607 b0092 b1061 b0094 b3933 b0093 b0089 b0759 b0758 b0082 b0087 b0081 b3972 b4041 b2699 b3832 b3650 b4483 b3651 b4040 b4039 b3813 b2683 b2682 b2896 b2897 b3344 b2412 b4169 b3934 b4042 b3702 b3701 b3463 b0084 b0083 b3462 b3464 b0095 b4018 b4346 b0091 b0088 b0085 b0090 b4170 b3825 b3652 b0097 b3348 b3965 b3465 b3466 b3470 b3826 b4166 b3096 | acpT birA cdd cpxA cysE ddlB dinI ftsA ftsN ftsQ ftsW galE galT mraW mraY mraZ murB plsB recA rmuC spoT tatD trmH ubiA ubiC uvrD ygaH ygaZ ygfX ygfY yheM zipA amiB cytR dgkA dnaA dnaN ftsE ftsI ftsL ftsX ftsY ftsZ iclR mcrB murC murD murE murG mutL pldB recG secM slyX trmA rsmD yhhL sirA yigL yjeS yqjB | holo-(acyl carrier protein) synthase 2 | bifunctional biotin-[acetylCoA carboxylase] holoenzyme synthetase/ DNA-binding transcriptional repressor, bio-5'-AMP-binding | cytidine/deoxycytidine deaminase | sensory histidine kinase in two-component regulatory system with CpxR | serine acetyltransferase | D-alanine:D-alanine ligase | DNA damage-inducible protein I | ATP-binding cell division protein involved in recruitment of FtsK to Z ring | essential cell division protein | membrane anchored protein involved in growth of wall at septum | integral membrane protein involved in stabilizing FstZ ring during cell division | UDP-galactose-4-epimerase | galactose-1-phosphate uridylyltransferase | S-adenosyl-dependent methyltransferase activity on membrane-located substrates | phospho-N-acetylmuramoyl-pentapeptide transferase | conserved protein | UDP-N-acetylenolpyruvoylglucosamine reductase, FAD-binding | glycerol-3-phosphate O-acyltransferase | DNA strand exchange and recombination protein with protease and nuclease activity | predicted recombination limiting protein | bifunctional (p)ppGpp synthetase II/ guanosine-3',5'-bis pyrophosphate 3'-pyrophosphohydrolase | DNase, magnesium-dependent | tRNA (Guanosine-2'-O-)-methyltransferase | p-hydroxybenzoate octaprenyltransferase | chorismate pyruvate lyase | DNA-dependent ATPase I and helicase II | predicted inner membrane protein | predicted transporter | predicted protein | predicted intraceullular sulfur oxidation protein | cell division protein involved in Z ring assembly | N-acetylmuramoyl-l-alanine amidase II | DNA-binding transcriptional dual regulator | diacylglycerol kinase | chromosomal replication initiator protein DnaA, DNA-binding transcriptional dual regulator | DNA polymerase III, beta subunit | predicted transporter subunit: ATP-binding component of ABC superfamily | transpeptidase involved in septal peptidoglycan synthesis (penicillin-binding protein 3) | membrane bound cell division protein at septum containing leucine zipper motif | predicted transporter subunit: membrane component of ABC superfamily | fused Signal Recognition Particle (SRP) receptor: membrane binding protein/conserved protein | GTP-binding tubulin-like cell division protein | DNA-binding transcriptional repressor | 5-methylcytosine-specific restriction enzyme McrBC, subunit McrB | UDP-N-acetylmuramate:L-alanine ligase | UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate:meso- diaminopimelate ligase | N-acetylglucosaminyl transferase | methyl-directed mismatch repair protein | lysophospholipase L(2) | ATP-dependent DNA helicase | regulator of secA translation | tRNA (uracil-5-)-methyltransferase | predicted methyltransferase | conserved inner membrane protein | conserved protein required for cell growth | predicted hydrolase | predicted Fe-S electron transport protein |
| 101 | 51 | 337 | -0.429 | 0.419 |  |  | 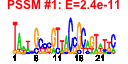 | 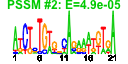 |  | b2220 b1412 b2078 b1413 b2075 b2076 b2077 b2045 b2043 b1366 b1535 b2177 b1736 b1738 b1737 b1734 b1733 b1735 b1201 b1579 b1845 b2218 b1456 b2044 b0913 b0920 b1063 b1163 b1181 b1325 b1327 b1438 b1501 b1769 b1771 b1772 b1773 b1774 b1775 b1776 b1931 b2067 b2178 b2179 b2180 b2623 b1368 b1365 b1436 b1815 b4538 | atoC azoR baeS hrpA mdtB mdtC mdtD wcaK wcaM ydaY ydeH yejA chbA chbB chbC chbF chbG chbR dhaR intQ ptrB rcsC rhsE wcaL ycaI ycbC yceB ycgF ycgN ycjG ycjY ydcQ ydeP ydjE ydjG ydjH ydjI ydjJ ydjK ydjL yedK yegE yejB yejE yejF yfjH ynaA ynaK yncJ yoaD yoeF | fused response regulator of ato operon, in two-component system with AtoS: response regulator/sigma54 interaction protein | NADH-azoreductase, FMN-dependent | sensory histidine kinase in two-component regulatory system with BaeR | ATP-dependent helicase | multidrug efflux system, subunit B | multidrug efflux system, subunit C | multidrug efflux system protein | predicted pyruvyl transferase | predicted colanic acid biosynthesis protein | conserved protein | predicted oligopeptide transporter subunit | N,N'-diacetylchitobiose-specific enzyme IIA component of PTS | N,N'-diacetylchitobiose-specific enzyme IIB component of PTS | N,N'-diacetylchitobiose-specific enzyme IIC component of PTS | cryptic phospho-beta-glucosidase, NAD(P)-binding | DNA-binding transcriptional dual regulator | predicted DNA-binding transcriptional regulator, dihydroxyacetone | protease II | hybrid sensory kinase in two-component regulatory system with RcsB and YojN | predicted glycosyl transferase | conserved inner membrane protein | predicted lipoprotein | predicted FAD-binding phosphodiesterase | L-Ala-D/L-Glu epimerase | predicted hydrolase | predicted DNA-binding transcriptional regulator | predicted oxidoreductase | predicted transporter | predicted kinase | predicted aldolase | predicted oxidoreductase, Zn-dependent and NAD(P)-binding | predicted protein | predicted diguanylate cyclase, GGDEF domain signalling protein | fused predicted oligopeptide transporter subunits of ABC superfamilly: ATP-binding components | CP4-57 prophage; predicted protein | Rac prophage; conserved protein | predicted phosphodiesterase |
| 95 | 49 | 336 | -0.373 | 0.398 |  |  | 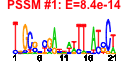 | 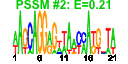 |  | b1591 b1532 b1069 b1638 b1912 b1456 b1662 b0995 b1202 b1769 b2249 b1590 b1762 b1606 b1529 b1663 b1399 b1400 b1812 b1636 b1363 b1610 b1960 b0916 b1206 b1220 b1289 b1322 b1320 b1464 b1491 b1528 b1577 b1601 b1605 b1624 b1659 b1657 b1694 b1688 b1695 b1770 b1813 b1835 b2006 b0835 b0952 b1816 b1811 | dmsD marB yceN pdxH pgsA rhsE ribC torR ycgV ydjE yfaY ynfH ynjI folM marC mdtK paaX paaY pabB pdxY trkG tus vsr ycaQ ychM ychO ycjD ycjF ycjW yddE yddW ydeA ydfE tqsA ydgI ydgJ ydhB ydhP ydiF ydiK ydiO ydjF nudL rsmF yeeW yliG ymbA yoaE yoaH | twin-argninine leader-binding protein for DmsA and TorA | predicted protein | predicted inner membrane protein | pyridoxine 5'-phosphate oxidase | phosphatidylglycerophosphate synthetase | riboflavin synthase, alpha subunit | DNA-binding response regulator in two-component regulatory system with TorS | predicted adhesin | predicted transporter | conserved protein | oxidoreductase, membrane subunit | dihydrofolate reductase isozyme | multidrug efflux system transporter | DNA-binding transcriptional repressor of phenylacetic acid degradation, aryl-CoA responsive | predicted hexapeptide repeat acetyltransferase | aminodeoxychorismate synthase, subunit I | pyridoxal kinase 2/pyridoxine kinase | Rac prophage; potassium transporter subunit | inhibitor of replication at Ter, DNA-binding protein | DNA mismatch endonuclease of very short patch repair | predicted invasin | conserved inner membrane protein | predicted DNA-binding transcriptional regulator | predicted liprotein | predicted arabinose transporter | predicted arginine/ornithine antiporter transporter | predicted oxidoreductase | fused predicted acetyl-CoA:acetoacetyl-CoA transferase: alpha subunit/beta subunit | predicted acyl-CoA dehydrogenase | predicted NUDIX hydrolase | predicted methyltransferase | predicted SAM-dependent methyltransferase | fused predicted membrane protein/conserved protein |
| 80 | 51 | 335 | -0.367 | 0.381 |  |  | 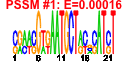 |  |  | b1270 b1343 b1334 b1606 b4426 b1529 b1663 b1333 b1271 b1322 b1320 b1340 b1341 b1342 b1344 b1414 b1464 b1491 b1624 b1639 b1659 b1640 b1655 b1657 b1688 b1752 b1781 b1834 b1835 b1871 b1523 b1525 b1526 b1753 b1754 b1756 b1816 b1731 b1562 b1702 b1564 b1563 b1321 b1496 b1565 b1703 b1791 b1828 b1868 b1332 b1843 | btuR dbpA fnr folM marC mdtK uspE yciK ycjF ycjW ydaL ydaM ydaN ttcA ydcF yddE yddW ydgJ ydhA ydhB anmK ydhO ydhP ydiK ydjZ yeaE yebT rsmF cmoB yneG yneI yneJ ynjA ynjB ynjD yoaE cedA hokD pps relB relE ycjX yddA ydfV ydiA yeaN yebQ yecE ynaJ yobB | cob(I)alamin adenolsyltransferase/cobinamide ATP-dependent adenolsyltransferase | ATP-dependent RNA helicase, specific for 23S rRNA | DNA-binding transcriptional dual regulator, global regulator of anaerobic growth | dihydrofolate reductase isozyme | predicted transporter | multidrug efflux system transporter | stress-induced protein | predicted oxoacyl-(acyl carrier protein) reductase, EmrKY-TolC system | conserved inner membrane protein | predicted DNA-binding transcriptional regulator | conserved protein | predicted diguanylate cyclase, GGDEF domain signalling protein | predicted Zn(II) transporter | predicted C32 tRNA thiolase | predicted liprotein | predicted oxidoreductase | predicted lipoprotein | predicted inner membrane protein | predicted methyltransferase | predicted S-adenosyl-L-methionine-dependent methyltransferase | predicted aldehyde dehydrogenase | predicted transporter subunit: ATP-binding component of ABC superfamily | fused predicted membrane protein/conserved protein | cell division modulator | Qin prophage; small toxic polypeptide | phosphoenolpyruvate synthase | Qin prophage; bifunctional antitoxin of the RelE-RelB toxin-antitoxin system/ transcriptional repressor | Qin prophage; toxin of the RelE-RelB toxin-antitoxin system | conserved protein with nucleoside triphosphate hydrolase domain | fused predicted multidrug transporter subunits of ABC superfamily: membrane component/ATP-binding component | Qin prophage; predicted protein |
| 50 | 54 | 339 | -0.346 | 0.435 |  |  |  |  |  | b1120 b1037 b0571 b0570 b1118 b1326 b1119 b1812 b1427 b1235 b1293 b1292 b1291 b1766 b0926 b0927 b1181 b1280 b1279 b1284 b1289 b1528 b1686 b1687 b1813 b1800 b1332 b1585 b1726 b1811 b1843 b1693 b1704 b1249 b0624 b1191 b1533 b1844 b1635 b1708 b1602 b1290 b1177 b1178 b1179 b1180 b1248 b1431 b1534 b1510 b1833 b1837 b1859 b1858 | cobB csgG cusR cusS lolE mpaA nagK pabB rimL rssB sapB sapC sapD sppA ycbK ycbL ycgN yciM yciS yciT ycjD ydeA ydiI ydiJ nudL yeaU ynaJ ynfC yniB yoaH yobB aroD aroH cls crcB cvrA eamA exoX gst nlpC pntB sapF ycgJ ycgK ycgL ycgM yciU ydcL ydeE ydeK yebS yebW znuB znuC | deacetylase of acetyl-CoA synthetase, NAD-dependent | outer membrane lipoprotein | DNA-binding response regulator in two-component regulatory system with CusS | sensory histidine kinase in two-component regulatory system with CusR, senses copper ions | outer membrane-specific lipoprotein transporter subunit | murein peptide amidase A | N-acetyl-D-glucosamine kinase | aminodeoxychorismate synthase, subunit I | ribosomal-protein-L7/L12-serine acetyltransferase | response regulator of RpoS | predicted antimicrobial peptide transporter subunit | protease IV (signal peptide peptidase) | conserved protein | predicted metal-binding enzyme | conserved inner membrane protein | predicted DNA-binding transcriptional regulator | predicted arabinose transporter | predicted FAD-linked oxidoreductase | predicted NUDIX hydrolase | predicted dehydrogenase | predicted inner membrane protein | predicted protein | 3-dehydroquinate dehydratase | 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase, tryptophan repressible | cardiolipin synthase 1 | predicted inner membrane protein associated with chromosome condensation | predicted cation/proton antiporter | cysteine and O-acetyl-L-serine efflux system | DNA exonuclease X | glutathionine S-transferase | predicted lipoprotein | pyridine nucleotide transhydrogenase, beta subunit | predicted isomerase/hydrolase | predicted transporter | zinc transporter subunit: membrane component of ABC superfamily | zinc transporter subunit: ATP-binding component of ABC superfamily |
| 84 | 57 | 332 | -0.326 | 0.441 |  |  | 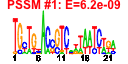 | 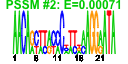 |  | b2697 b2480 b0437 b0623 b2478 b3860 b0529 b2479 b3385 b4480 b2514 b0178 b4000 b0439 b0179 b2477 b0134 b0133 b0131 b0143 b2926 b4226 b2217 b2216 b3386 b2914 b0120 b0121 b0177 b0424 b0426 b0490 b3764 b4243 b4394 b3149 b3190 b4395 b0438 b3357 b2925 b4259 b2215 b0425 b4260 b3622 b3308 b2185 b3300 b0687 b2793 b3919 b4258 b0488 b0489 b0491 b2794 | alaS bcp clpP cspE dapA dsbA folD gcvR gph hdfR hisS skp hupA lon lpxD nlpB panB panC panD pcnB pgk ppa rcsB rcsD rpe rpiA speD speE yaeT yajL yajQ ybbL yifE yjgF yjjX diaA yrbA ytjC clpX crp fbaA holC ompC panE pepA rfaL rplE rplY secY seqA syd tpiA valS ybbJ qmcA ybbM queF | alanyl-tRNA synthetase | thiol peroxidase, thioredoxin-dependent | proteolytic subunit of ClpA-ClpP and ClpX-ClpP ATP-dependent serine proteases | DNA-binding transcriptional repressor | dihydrodipicolinate synthase | periplasmic protein disulfide isomerase I | bifunctional 5,10-methylene-tetrahydrofolate dehydrogenase/ 5,10-methylene-tetrahydrofolate cyclohydrolase | DNA-binding transcriptional repressor, regulatory protein accessory to GcvA | phosphoglycolate phosphatase | DNA-binding transcriptional regulator | histidyl tRNA synthetase | periplasmic chaperone | HU, DNA-binding transcriptional regulator, alpha subunit | DNA-binding ATP-dependent protease La | UDP-3-O-(3-hydroxymyristoyl)-glucosamine N-acyltransferase | lipoprotein | 3-methyl-2-oxobutanoate hydroxymethyltransferase | pantothenate synthetase | aspartate 1-decarboxylase | poly(A) polymerase I | phosphoglycerate kinase | inorganic pyrophosphatase | DNA-binding response regulator in two-component regulatory system with RcsC and YojN | phosphotransfer intermediate protein in two-component regulatory system with RcsBC | D-ribulose-5-phosphate 3-epimerase | ribose 5-phosphate isomerase, constitutive | S-adenosylmethionine decarboxylase | spermidine synthase (putrescine aminopropyltransferase) | conserved protein | predicted nucleotide binding protein | predicted transporter subunit: ATP-binding component of ABC superfamily | ketoacid-binding protein | thiamin metabolism associated protein | DnaA initiator-associating factor for replication initiation | predicted DNA-binding transcriptional regulator | phosphoglyceromutase 2, co-factor independent | ATPase and specificity subunit of ClpX-ClpP ATP-dependent serine protease | DNA-binding transcriptional dual regulator | fructose-bisphosphate aldolase, class II | DNA polymerase III, chi subunit | outer membrane porin protein C | 2-dehydropantoate reductase, NADPH-specific | aminopeptidase A, a cyteinylglycinase | O-antigen ligase | 50S ribosomal subunit protein L5 | 50S ribosomal subunit protein L25 | preprotein translocase membrane subunit | regulatory protein for replication initiation | predicted protein | triosephosphate isomerase | valyl-tRNA synthetase | conserved inner membrane protein | predicted protease, membrane anchored | predicted inner membrane protein |
| 6 | 51 | 332 | -0.306 | 0.405 |  |  | 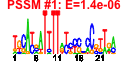 | 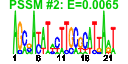 |  | b2213 b1453 b1533 b1384 b0654 b3174 b2469 b1702 b1494 b0813 b2176 b2170 b1616 b0844 b0845 b0901 b0899 b0900 b0916 b0992 b1058 b1121 b1242 b1439 b1447 b1496 b1495 b1534 b1565 b1601 b1644 b1645 b1791 b1828 b1959 b1978 b2098 b2099 b2100 b2178 b2179 b2180 b2246 b2247 b2248 b1172 b1582 b1755 b0968 b1355 b1790 | ada ansP eamA feaR gltJ leuU narQ pps pqqL rhtA rtn setB uidB ybjI ybjJ ycaK ycaM ycaN ycaQ yccM yceO ycfZ ychE ydcR ydcZ yddA yddB ydeE ydfV tqsA ydhJ ydhK yeaN yebQ yedA yeeJ yegT yegU yegV yejB yejE yejF yfaV yfaW yfaX ymgG ynfA ynjC yccX ydaG yeaM | fused DNA-binding transcriptional dual regulator/O6-methylguanine-DNA methyltransferase | L-asparagine transporter | cysteine and O-acetyl-L-serine efflux system | DNA-binding transcriptional dual regulator | glutamate and aspartate transporter subunit | tRNA-Leu | sensory histidine kinase in two-component regulatory system with NarP (NarL) | phosphoenolpyruvate synthase | predicted peptidase | threonine and homoserine efflux system | conserved protein | lactose/glucose efflux system | predicted transporter | predicted DNA-binding transcriptional regulator | predicted 4Fe-4S membrane protein | predicted protein | predicted inner membrane protein | fused predicted DNA-binding transcriptional regulator/predicted amino transferase | fused predicted multidrug transporter subunits of ABC superfamily: membrane component/ATP-binding component | predicted porin protein | Qin prophage; predicted protein | undecaprenyl pyrophosphate phosphatase | conserved inner membrane protein | adhesin | predicted nucleoside transporter | predicted hydrolase | predicted kinase | predicted oligopeptide transporter subunit | fused predicted oligopeptide transporter subunits of ABC superfamilly: ATP-binding components | predicted enolase | fused transporter subunits of ABC superfamily: membrane components | predicted acylphosphatase | Rac prophage; predicted protein |
| 68 | 51 | 353 | -0.287 | 0.402 |  |  |  |  |  | b1481 b1218 b0566 b1990 b1516 b1362 b1519 b0994 b1197 b0662 b0608 b0843 b0920 b1327 b1357 b1359 b1360 b1361 b1406 b1598 b1696 b1956 b2067 b0837 b4525 b1217 b0575 b0574 b0855 b0856 b0857 b1838 b1296 b0993 b0788 b0806 b0898 b0901 b0900 b1085 b1328 b1428 b1432 b1490 b1536 b1537 b1542 b1648 b1668 b1800 b1454 | bdm chaC envY erfK lsrB rzpR tam torT treA ubiF ybdR ybjH ycbC ycjY ydaS ydaU ydaV ydaW ydbC ydgD ydiP yedQ yegE yliI ymjC chaB cusA cusB potG potH potI pphA puuP torS ybhN ybiM ycaD ycaK ycaN yceQ ycjZ ydcK ydcM yddV ydeI ydeJ ydfI ydhL ydhS yeaU yncG | biofilm-dependent modulation protein | regulatory protein for cation transport | DNA-binding transcriptional activator of porin biosynthesis | conserved protein with NAD(P)-binding Rossmann-fold domain | AI2 transporter | trans-aconitate methyltransferase | periplasmic sensory protein associated with the TorRS two-component regulatory system | periplasmic trehalase | 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol oxygenase | predicted oxidoreductase, Zn-dependent and NAD(P)-binding | predicted protein | conserved inner membrane protein | predicted hydrolase | Rac prophage; predicted DNA-binding transcriptional regulator | Rac prophage; conserved protein | Rac prophage; predicted DNA replication protein | predicted oxidoreductase, NAD(P)-binding | predicted peptidase | predicted DNA-binding transcriptional regulator | predicted diguanylate cyclase | predicted diguanylate cyclase, GGDEF domain signalling protein | predicted dehydrogenase | predicted cation regulator | copper/silver efflux system, membrane component | copper/silver efflux system, membrane fusion protein | putrescine transporter subunit: ATP-binding component of ABC superfamily | putrescine transporter subunit: membrane component of ABC superfamily | serine/threonine-specific protein phosphatase 1 | putrescine importer | hybrid sensory histidine kinase in two-component regulatory system with TorR | predicted transporter | conserved protein | predicted enzyme | predicted transposase | predicted mannonate dehydrogenase | conserved protein with FAD/NAD(P)-binding domain |
| 129 | 67 | 328 | -0.280 | 0.399 |  |  |  |  |  | b1193 b1187 b1604 b2064 b2132 b1270 b2198 b1120 b1991 b1874 b1418 b1961 b1215 b1192 b1117 b1118 b1622 b1819 b1424 b1069 b1119 b1222 b1186 b1912 b1768 b1658 b1281 b1662 b1272 b1059 b1584 b1766 b1429 b1430 b1274 b1324 b2232 b1035 b1219 b1271 b1280 b1268 b1279 b1284 b1383 b1419 b1654 b1655 b1778 b1777 b1780 b1787 b1794 b1839 b1908 b1875 b1869 b1870 b1871 b2249 b1382 b1582 b1583 b1590 b1762 b1809 b1820 | emtA fadR ydgH asmA bglX btuR ccmD cobB cobT cutC cybB dcm kdsA ldcA lolD lolE malY manZ mdoD yceN nagK narX nhaB pgsA pncA purR pyrF ribC sohB solA speG sppA tehA tehB topA tpx ubiG ycdY ychN yciK yciM yciQ yciS yciT ydbL ydcA grxD ydhO msrB yeaC yeaD yeaK yeaP yebY yecA yecM yecN cmoA cmoB yfaY ynbE ynfA ynfB ynfH ynjI yoaB yobD | lytic murein endotransglycosylase E | DNA-binding transcriptional dual regulator of fatty acid metabolism | predicted protein | predicted assembly protein | beta-D-glucoside glucohydrolase, periplasmic | cob(I)alamin adenolsyltransferase/cobinamide ATP-dependent adenolsyltransferase | cytochrome c biogenesis protein | deacetylase of acetyl-CoA synthetase, NAD-dependent | nicotinate-nucleotide dimethylbenzimidazole-P phophoribosyl transferase | copper homeostasis protein | cytochrome b561 | DNA cytosine methylase | 3-deoxy-D-manno-octulosonate 8-phosphate synthase | L,D-carboxypeptidase A | outer membrane-specific lipoprotein transporter subunit | bifunctional beta-cystathionase, PLP-dependent/ regulator of maltose regulon | mannose-specific enzyme IID component of PTS | glucan biosynthesis protein, periplasmic | predicted inner membrane protein | N-acetyl-D-glucosamine kinase | sensory histidine kinase in two-component regulatory system with NarL | sodium:proton antiporter | phosphatidylglycerophosphate synthetase | nicotinamidase/pyrazinamidase | DNA-binding transcriptional repressor, hypoxanthine-binding | orotidine-5'-phosphate decarboxylase | riboflavin synthase, alpha subunit | predicted inner membrane peptidase | N-methyltryptophan oxidase, FAD-binding | spermidine N1-acetyltransferase | protease IV (signal peptide peptidase) | potassium-tellurite ethidium and proflavin transporter | predicted S-adenosyl-L-methionine-dependent methyltransferase | DNA topoisomerase I, omega subunit | lipid hydroperoxide peroxidase | bifunctional 3-demethylubiquinone-9 3-methyltransferase/ 2-octaprenyl-6-hydroxy phenol methylase | conserved protein | predicted oxoacyl-(acyl carrier protein) reductase, EmrKY-TolC system | conserved inner membrane protein | predicted DNA-binding transcriptional regulator | predicted lipoprotein | methionine sulfoxide reductase B | predicted diguanylate cyclase | conserved metal-binding protein | predicted metal-binding enzyme | predicted methyltransferase | oxidoreductase, membrane subunit |
| 142 | 59 | 336 | -0.268 | 0.415 |  |  |  |  |  | b1876 b1711 b2413 b2065 b1591 b1185 b1193 b1187 b1099 b1855 b1048 b1049 b1650 b1096 b0688 b1126 b1125 b1124 b1123 b1204 b1131 b1286 b1804 b1652 b1822 b1628 b1629 b1630 b1632 b1631 b1294 b1293 b1292 b1291 b1238 b1098 b1637 b2066 b1749 b0686 b0819 b0820 b0969 b0967 b1097 b1203 b1722 b1806 b1807 b1856 b2171 b2186 b1134 b1135 b1295 b1523 b1524 b1727 b1857 | argS btuC cysZ dcd dmsD dsbB emtA fadR holB lpxM mdoG mdoH nemA pabC pgm potA potB potC potD pth purB rnb rnd rnt rrmA rsxB rsxC rsxD rsxE rsxG sapA sapB sapC sapD tdk tmk tyrS udk xthA ybfF ybiS ybiT yccK yccW yceG ychF ydiY yeaY yeaZ yebA yeiP yejK nudJ rluE ymjA yneG yneH yniC znuA | arginyl-tRNA synthetase | vitamin B12 transporter subunit: membrane component of ABC superfamily | predicted inner membrane protein | 2'-deoxycytidine 5'-triphosphate deaminase | twin-argninine leader-binding protein for DmsA and TorA | oxidoreductase that catalyzes reoxidation of DsbA protein disulfide isomerase I | lytic murein endotransglycosylase E | DNA-binding transcriptional dual regulator of fatty acid metabolism | DNA polymerase III, delta prime subunit | myristoyl-acyl carrier protein (ACP)-dependent acyltransferase | glucan biosynthesis protein, periplasmic | glucan biosynthesis: glycosyl transferase | N-ethylmaleimide reductase, FMN-linked | 4-amino-4-deoxychorismate lyase component of para-aminobenzoate synthase multienzyme complex | phosphoglucomutase | polyamine transporter subunit | peptidyl-tRNA hydrolase | adenylosuccinate lyase | ribonuclease II | ribonuclease D | ribonuclease T (RNase T) | 23S rRNA m1G745 methyltransferase | predicted iron-sulfur protein | fused predicted 4Fe-4S ferredoxin-type protein/conserved protein | predicted inner membrane oxidoreductase | predicted inner membrane NADH-quinone reductase | predicted oxidoreductase | predicted antimicrobial peptide transporter subunit | thymidine kinase/deoxyuridine kinase | thymidylate kinase | tyrosyl-tRNA synthetase | uridine/cytidine kinase | exonuclease III | conserved protein | fused predicted transporter subunits of ABC superfamily: ATP-binding components | predicted sulfite reductase subunit | predicted methyltransferase | predicted aminodeoxychorismate lyase | predicted GTP-binding protein | predicted lipoprotein | predicted peptidase | predicted elongtion factor | nucleotide associated protein | bifunctional thiamin pyrimidine pyrophosphate hydrolase/ thiamin pyrophosphate hydrolase | 23S rRNA pseudouridine synthase | predicted protein | predicted glutaminase | predicted hydrolase | zinc transporter subunit: periplasmic-binding component of ABC superfamily |
| 20 | 62 | 331 | -0.214 | 0.422 |  |  |  | 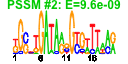 |  | b4015 b4014 b4016 b4187 b3959 b3958 b3359 b2664 b3544 b3540 b2663 b2976 b2979 b4467 b3870 b1285 b3770 b0220 b3661 b2742 b2676 b2673 b1897 b4107 b0953 b2741 b2465 b3519 b1004 b0219 b0325 b0329 b0453 b0897 b1003 b1783 b1795 b2112 b2602 b2638 b2660 b2809 b2975 b2989 b3000 b3073 b4199 b4329 b1998 b2543 b3830 b3960 b2601 b3543 b3542 b3541 b2574 b3089 b2600 b0328 b0906 b4330 | aceA aceB aceK aidB argB argC argD csiR dppA dppF gabP glcB glcD glcF glnA gmr ilvE ivy nlpA nlpD nrdF nrdH otsB yjdN rmf rpoS tktB treF wrbA yafV yahK yahO ybaY ycaC yccJ yeaG yeaQ yehE yfiL yfjU ygaF ygdI glcA yghU ygjG yjfY yjiG yphA ysgA argH aroF dppB dppC dppD nadB sstT tyrA yahN ycaP yjiH | isocitrate lyase | malate synthase A | isocitrate dehydrogenase kinase/phosphatase | isovaleryl CoA dehydrogenase | acetylglutamate kinase | N-acetyl-gamma-glutamylphosphate reductase, NAD(P)-binding | bifunctional acetylornithine aminotransferase/ succinyldiaminopimelate aminotransferase | DNA-binding transcriptional dual regulator | dipeptide transporter | gamma-aminobutyrate transporter | malate synthase G | glycolate oxidase subunit, FAD-linked | glycolate oxidase iron-sulfur subunit | glutamine synthetase | modulator of Rnase II stability | branched-chain amino-acid aminotransferase | inhibitor of vertebrate C-lysozyme | cytoplasmic membrane lipoprotein-28 | predicted outer membrane lipoprotein | ribonucleoside-diphosphate reductase 2, beta subunit, ferritin-like protein | glutaredoxin-like protein | trehalose-6-phosphate phosphatase, biosynthetic | conserved protein | ribosome modulation factor | RNA polymerase, sigma S (sigma 38) factor | transketolase 2, thiamin-binding | cytoplasmic trehalase | predicted flavoprotein in Trp regulation | predicted C-N hydrolase family amidase, NAD(P)-binding | predicted oxidoreductase, Zn-dependent and NAD(P)-binding | predicted protein | predicted hydrolase | conserved protein with nucleoside triphosphate hydrolase domain | conserved inner membrane protein | predicted enzyme | glycolate transporter | predicted S-transferase | putrescine:2-oxoglutaric acid aminotransferase, PLP-dependent | predicted inner membrane protein | argininosuccinate lyase | 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase, tyrosine-repressible | quinolinate synthase, L-aspartate oxidase (B protein) subunit | sodium:serine/threonine symporter | fused chorismate mutase T/prephenate dehydrogenase | neutral amino-acid efflux system |
| 139 | 68 | 335 | -0.181 | 0.429 |  |  |  |  |  | b1876 b2413 b1185 b1099 b1855 b1096 b1126 b1125 b1124 b1123 b1131 b1804 b1652 b1822 b1628 b1629 b1098 b1749 b0820 b0969 b0967 b1097 b1806 b1807 b1856 b2186 b1134 b1135 b1524 b1857 b1767 b0930 b1866 b0526 b2140 b0954 b2400 b0477 b2231 b0524 b0915 b0914 b1633 b1865 b0525 b0951 b0393 b1627 b0859 b1863 b1763 b1133 b0766 b0807 b0955 b0961 b0960 b1233 b1253 b1254 b1255 b1434 b1435 b1864 b1867 b2081 b2325 b1808 | argS cysZ dsbB holB lpxM pabC potA potB potC potD purB rnd rnt rrmA rsxB rsxC tmk xthA ybiT yccK yccW yceG yeaY yeaZ yebA yejK nudJ rluE yneH znuA ansA asnS aspS cysS dusC fabA gltX gsk gyrA lpxH lpxK msbA nth nudB ppiB pqiB rdgC rsxA rumB ruvC topB mnmA ybhA ybiN ycbZ yccF yccS ychJ yciA yciB yciC ydcN ydcP yebC yecD yegQ yfcL yoaA | arginyl-tRNA synthetase | predicted inner membrane protein | oxidoreductase that catalyzes reoxidation of DsbA protein disulfide isomerase I | DNA polymerase III, delta prime subunit | myristoyl-acyl carrier protein (ACP)-dependent acyltransferase | 4-amino-4-deoxychorismate lyase component of para-aminobenzoate synthase multienzyme complex | polyamine transporter subunit | adenylosuccinate lyase | ribonuclease D | ribonuclease T (RNase T) | 23S rRNA m1G745 methyltransferase | predicted iron-sulfur protein | fused predicted 4Fe-4S ferredoxin-type protein/conserved protein | thymidylate kinase | exonuclease III | fused predicted transporter subunits of ABC superfamily: ATP-binding components | predicted sulfite reductase subunit | predicted methyltransferase | predicted aminodeoxychorismate lyase | predicted lipoprotein | predicted peptidase | nucleotide associated protein | bifunctional thiamin pyrimidine pyrophosphate hydrolase/ thiamin pyrophosphate hydrolase | 23S rRNA pseudouridine synthase | predicted glutaminase | zinc transporter subunit: periplasmic-binding component of ABC superfamily | cytoplasmic L-asparaginase I | asparaginyl tRNA synthetase | aspartyl-tRNA synthetase | cysteinyl-tRNA synthetase | tRNA-dihydrouridine synthase C | beta-hydroxydecanoyl thioester dehydrase | glutamyl-tRNA synthetase | inosine/guanosine kinase | DNA gyrase (type II topoisomerase), subunit A | UDP-2,3-diacylglucosamine pyrophosphatase | lipid A 4'kinase | fused lipid transporter subunits of ABC superfamily: membrane component/ATP-binding component | DNA glycosylase and apyrimidinic (AP) lyase (endonuclease III) | dATP pyrophosphohydrolase | peptidyl-prolyl cis-trans isomerase B (rotamase B) | paraquat-inducible protein B | DNA-binding protein, non-specific | predicted inner membrane subunit | 23S rRNA m(5)U747 methyltransferase | component of RuvABC resolvasome, endonuclease | DNA topoisomerase III | tRNA (5-methylaminomethyl-2-thiouridylate)-methyltransferase | predicted hydrolase | predicted SAM-dependent methyltransferase | conserved inner membrane protein | conserved protein | predicted DNA-binding transcriptional regulator | predicted protein | conserved protein with nucleoside triphosphate hydrolase domain |
| 102 | 51 | 346 | -0.168 | 0.466 |  |  |  |  |  | b2222 b2224 b2221 b2223 b2364 b2538 b2537 b2481 b2482 b2483 b2484 b2486 b2487 b2488 b2491 b2406 b1423 b1689 b2333 b2334 b2335 b2336 b1122 b1527 b2544 b2468 b2715 b2220 b2366 b2365 b1879 b2492 b2789 b2541 b2540 b2542 b2539 b2373 b2046 b1121 b1470 b2072 b2073 b2292 b2298 b2337 b2372 b2467 b2447 b2999 b2382 | atoA atoB atoD atoE dsdC hcaE hcaR hyfA hyfB hyfC hyfD hyfF hyfG hyfH hyfR xapB ydcJ ydiL yfcP yfcQ yfcR yfcS ymfA yneK yphB aegA ascF atoC dsdA dsdX flhA focB gudP hcaB hcaC hcaD hcaF oxc wzxC ycfZ yddJ yegK yegL yfbS yfcC yfdV nudK yffP ypdC | acetyl-CoA:acetoacetyl-CoA transferase, beta subunit | acetyl-CoA acetyltransferase | acetyl-CoA:acetoacetyl-CoA transferase, alpha subunit | short chain fatty acid transporter | DNA-binding transcriptional dual regulator | 3-phenylpropionate dioxygenase, large (alpha) subunit | DNA-binding transcriptional activator of 3-phenylpropionic acid catabolism | hydrogenase 4, 4Fe-4S subunit | hydrogenase 4, membrane subunit | hydrogenase 4, subunit | hydrogenase 4, Fe-S subunit | DNA-binding transcriptional activator, formate sensing | xanthosine transporter | conserved protein | predicted fimbrial-like adhesin protein | predicted periplasmic pilus chaperone | predicted inner membrane protein | predicted protein | fused predicted oxidoreductase: FeS binding subunit/NAD/FAD-binding subunit | fused cellobiose/arbutin/salicin-specific PTS enzymes: IIB component/IC component | fused response regulator of ato operon, in two-component system with AtoS: response regulator/sigma54 interaction protein | D-serine ammonia-lyase | predicted transporter | predicted flagellar export pore protein | predicted formate transporter | predicted D-glucarate transporter | 2,3-dihydroxy-2,3-dihydrophenylpropionate dehydrogenase | 3-phenylpropionate dioxygenase, predicted ferredoxin subunit | phenylpropionate dioxygenase, ferredoxin reductase subunit | 3-phenylpropionate dioxygenase, small (beta) subunit | predicted oxalyl-CoA decarboxylase | colanic acid exporter | predicted NUDIX hydrolase | CPZ-55 prophage; predicted protein | predicted DNA-binding protein |
| 146 | 68 | 339 | -0.110 | 0.393 |  |  | 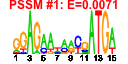 |  |  | b1685 b4015 b4014 b1927 b0435 b1000 b0999 b0484 b2664 b0487 b2097 b2661 b2663 b2662 b1492 b3447 b1967 b2127 b1051 b2742 b1283 b1482 b1739 b4376 b1896 b1723 b0871 b0384 b0953 b2741 b4459 b1480 b3453 b0325 b0419 b0485 b0486 b0753 b0773 b0767 b0789 b0790 b0803 b1164 b1724 b1836 b1970 b2080 b2086 b2112 b2638 b2659 b2665 b3003 b3000 b3073 b3153 b3448 b4377 b4378 b4518 b1165 b1166 b1449 b1450 b1725 b2137 b4568 | ydiH aceA aceB amyA bolA cbpA cbpM copA csiR cueR fbaB gabD gabP gabT gadC ggt hchA mlrA msyB nlpD osmB osmC osmE osmY otsA pfkB poxB psiF rmf rpoS sra ugpB yahK yajO ybaS ybaT ybgS ybhB pgl ybhO ybhP ybiI ycgZ ydiZ yebV yedX yegP yegS yehE yfjU csiD ygaU yghA ygjG yhbO yhhA yjjU yjjV ymdF ymgA ymgB yncB yncC yniA yohF ytjA | predicted protein | isocitrate lyase | malate synthase A | cytoplasmic alpha-amylase | regulator of penicillin binding proteins and beta lactamase transcription (morphogene) | curved DNA-binding protein, DnaJ homologue that functions as a co-chaperone of DnaK | modulator of CbpA co-chaperone | copper transporter | DNA-binding transcriptional dual regulator | DNA-binding transcriptional activator of copper-responsive regulon genes | fructose-bisphosphate aldolase class I | succinate-semialdehyde dehydrogenase I, NADP-dependent | gamma-aminobutyrate transporter | 4-aminobutyrate aminotransferase, PLP-dependent | predicted glutamate:gamma-aminobutyric acid antiporter | gamma-glutamyltranspeptidase | Hsp31 molecular chaperone | DNA-binding transcriptional regulator | predicted outer membrane lipoprotein | lipoprotein | osmotically inducible, stress-inducible membrane protein | DNA-binding transcriptional activator | periplasmic protein | trehalose-6-phosphate synthase | 6-phosphofructokinase II | pyruvate dehydrogenase (pyruvate oxidase), thiamin-dependent, FAD-binding | conserved protein | ribosome modulation factor | RNA polymerase, sigma S (sigma 38) factor | 30S ribosomal subunit protein S22 | glycerol-3-phosphate transporter subunit | predicted oxidoreductase, Zn-dependent and NAD(P)-binding | 2-carboxybenzaldehyde reductase, function unknown | predicted glutaminase | predicted transporter | predicted kinase inhibitor | 6-phosphogluconolactonase | cardiolipin synthase 2 | predicted DNase | phosphatidylglycerol kinase, metal-dependent | predicted glutathionylspermidine synthase, with NAD(P)-binding Rossmann-fold domain | putrescine:2-oxoglutaric acid aminotransferase, PLP-dependent | predicted intracellular protease | predicted esterase | predicted DNA-binding transcriptional regulator | predicted phosphotransferase/kinase | predicted oxidoreductase with NAD(P)-binding Rossmann-fold domain |
| 87 | 63 | 324 | -0.091 | 0.442 |  |  | 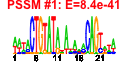 | 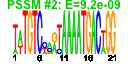 |  | b0063 b0061 b2841 b1901 b0396 b4131 b4132 b3556 b1557 b1558 b0990 b0989 b1552 b0231 b3645 b0799 b2943 b2706 b1140 b4043 b0113 b0060 b2616 b2698 b1861 b1860 b2704 b0958 b1184 b1183 b4058 b0779 b0233 b0234 b1457 b1728 b1741 b1848 b2840 b1141 b1142 b1144 b1147 b1148 b1146 b1458 b0062 b1900 b4460 b4133 b4044 b1061 b2699 b2009 b2702 b2705 b2703 b4059 b3813 b0232 b1847 b4347 b1149 | araB araD araE araF araJ cadA cadB cspA cspB cspF cspG cspH cspI dinB dinD dinG galP gutM intE lexA pdhR polB recN recX ruvA ruvB srlB sulA umuC umuD uvrA uvrB yafO yafP ydcD ydjM cho yebG ygeA xisE ymfJ ymfL ymfM ymfT araA araG araH cadC dinF dinI recA sbmC srlA srlD srlE ssb uvrD yafN yebF yjiW ymfN | L-ribulokinase | L-ribulose-5-phosphate 4-epimerase | arabinose transporter | L-arabinose transporter subunit | predicted transporter | lysine decarboxylase 1 | predicted lysine/cadaverine transporter | major cold shock protein | Qin prophage; cold shock protein | DNA-binding transcriptional regulator | stress protein, member of the CspA-family | DNA polymerase IV | DNA-damage-inducible protein | ATP-dependent DNA helicase | D-galactose transporter | DNA-binding transcriptional activator of glucitol operon | e14 prophage; predicted integrase | DNA-binding transcriptional repressor of SOS regulon | DNA-binding transcriptional dual regulator | DNA polymerase II | recombination and repair protein | regulatory protein for RecA | component of RuvABC resolvasome, regulatory subunit | ATP-dependent DNA helicase, component of RuvABC resolvasome | glucitol/sorbitol-specific enzyme IIA component of PTS | SOS cell division inhibitor | DNA polymerase V, subunit C | DNA polymerase V, subunit D | ATPase and DNA damage recognition protein of nucleotide excision repair excinuclease UvrABC | excinulease of nucleotide excision repair, DNA damage recognition component | predicted toxin of the YafO-YafN toxin-antitoxin system | predicted acyltransferase with acyl-CoA N-acyltransferase domain | predicted protein | predicted inner membrane protein regulated by LexA | endonuclease of nucleotide excision repair | conserved protein regulated by LexA | predicted racemase | e14 prophage; predicted excisionase | e14 prophage; predicted protein | e14 prophage; predicted DNA-binding transcriptional regulator | L-arabinose isomerase | fused L-arabinose transporter subunits of ABC superfamily: ATP-binding components | fused L-arabinose transporter subunits of ABC superfamily: membrane components | DNA-binding transcriptional activator | DNA-damage-inducible SOS response protein | DNA damage-inducible protein I | DNA strand exchange and recombination protein with protease and nuclease activity | DNA gyrase inhibitor | glucitol/sorbitol-specific enzyme IIC component of PTS | sorbitol-6-phosphate dehydrogenase | glucitol/sorbitol-specific enzyme IIB component of PTS | single-stranded DNA-binding protein | DNA-dependent ATPase I and helicase II | predicted antitoxin of the YafO-YafN toxin-antitoxin system | conserved protein |
| 133 | 44 | 342 | -0.085 | 0.446 |  |  | 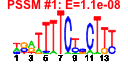 | 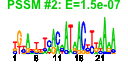 |  | b0715 b2212 b0875 b0619 b0617 b0616 b0613 b0614 b0123 b0554 b0229 b0873 b0872 b0878 b0879 b1065 b0149 b0692 b0693 b2103 b0994 b2311 b0005 b0267 b0282 b0283 b0286 b0404 b0496 b0555 b0608 b1700 b1730 b1789 b1862 b1985 b2003 b2004 b2437 b2740 b0515 b0837 b1788 b2390 | abrB alkB aqpZ dpiB citD citE citG citX cueO essD lfhA hcp hcr macA macB mdtH mrcB potE speF thiD torT ubiX yaaX yagA yagP yagQ yagT acpH ybbP ybcS ybdR ydiT ydjO yeaL yebB yeeO yeeT yeeU eutR ygbN ylbA yliI yoaI ypeC | predicted regulator | oxidative demethylase of N1-methyladenine or N3-methylcytosine DNA lesions | aquaporin | sensory histidine kinase in two-component regulatory system with citB | citrate lyase, acyl carrier (gamma) subunit | citrate lyase, citryl-ACP lyase (beta) subunit | triphosphoribosyl-dephospho-CoA transferase | apo-citrate lyase phosphoribosyl-dephospho-CoA transferase | multicopper oxidase (laccase) | DLP12 prophage; predicted phage lysis protein | hybrid-cluster [4Fe-2S-2O] protein in anaerobic terminal reductases | HCP oxidoreductase, NADH-dependent | macrolide transporter subunit, membrane fusion protein (MFP) component | fused macrolide transporter subunits of ABC superfamily: ATP-binding component/membrane component | predicted drug efflux system | fused glycosyl transferase and transpeptidase | putrescine/proton symporter: putrescine/ornithine antiporter | ornithine decarboxylase isozyme, inducible | bifunctional hydroxy-methylpyrimidine kinase/ hydroxy-phosphomethylpyrimidine kinase | periplasmic sensory protein associated with the TorRS two-component regulatory system | 3-octaprenyl-4-hydroxybenzoate carboxy-lyase | predicted protein | CP4-6 prophage; predicted DNA-binding transcriptional regulator | predicted transcriptional regulator | conserved protein | predicted xanthine dehydrogenase, 2Fe-2S subunit | predicted inner membrane protein | DLP12 prophage; predicted lysozyme | predicted oxidoreductase, Zn-dependent and NAD(P)-binding | predicted 4Fe-4S ferredoxin-type protein | conserved inner membrane protein | predicted multidrug efflux system | CP4-44 prophage; predicted protein | CP4-44 prophage; antitoxin of the YeeV-YeeU toxin-antitoxin system | predicted DNA-binding transcriptional regulator | predicted transporter | predicted dehydrogenase |
| 35 | 62 | 339 | -0.084 | 0.409 |  |  | 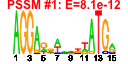 | 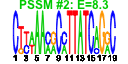 |  | b3336 b0435 b0487 b1492 b3049 b1967 b2924 b1283 b1739 b4376 b1723 b0871 b0384 b4459 b1480 b2464 b0419 b0643 b0806 b0865 b0867 b1164 b1724 b2086 b2672 b2665 b3153 b3362 b4057 b4377 b4378 b4518 b1165 b1166 b1586 b1725 b1957 b4568 b4187 b3012 b0220 b1897 b4107 b2465 b3519 b1004 b0219 b0329 b0453 b0866 b0897 b1003 b1783 b1795 b2602 b2660 b2809 b3491 b4056 b4199 b2543 b3830 | bfr bolA cueR gadC glgS hchA mscS osmB osmE osmY pfkB poxB psiF sra talA yajO ybeL ybiM ybjP ybjR ycgZ ydiZ yegS ygaM ygaU yhbO yhfG yjbR yjjU yjjV ymdF ymgA ymgB ynfD yniA yodC ytjA aidB dkgA ivy otsB yjdN tktB treF wrbA yafV yahO ybaY ybjQ ycaC yccJ yeaG yeaQ yfiL ygaF ygdI yhiM yjbQ yjfY yphA ysgA | bacterioferritin, iron storage and detoxification protein | regulator of penicillin binding proteins and beta lactamase transcription (morphogene) | DNA-binding transcriptional activator of copper-responsive regulon genes | predicted glutamate:gamma-aminobutyric acid antiporter | predicted glycogen synthesis protein | Hsp31 molecular chaperone | mechanosensitive channel | lipoprotein | DNA-binding transcriptional activator | periplasmic protein | 6-phosphofructokinase II | pyruvate dehydrogenase (pyruvate oxidase), thiamin-dependent, FAD-binding | conserved protein | 30S ribosomal subunit protein S22 | transaldolase A | 2-carboxybenzaldehyde reductase, function unknown | predicted protein | predicted lipoprotein | predicted amidase and lipoprotein | phosphatidylglycerol kinase, metal-dependent | predicted intracellular protease | predicted esterase | predicted DNase | predicted phosphotransferase/kinase | isovaleryl CoA dehydrogenase | 2,5-diketo-D-gluconate reductase A | inhibitor of vertebrate C-lysozyme | trehalose-6-phosphate phosphatase, biosynthetic | transketolase 2, thiamin-binding | cytoplasmic trehalase | predicted flavoprotein in Trp regulation | predicted C-N hydrolase family amidase, NAD(P)-binding | predicted outer membrane lipoprotein | predicted hydrolase | conserved protein with nucleoside triphosphate hydrolase domain | conserved inner membrane protein | predicted enzyme | predicted inner membrane protein |
| 108 | 62 | 337 | -0.068 | 0.450 |  |  | 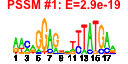 | 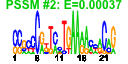 |  | b1597 b2079 b0811 b0655 b0962 b2074 b1014 b1015 b1301 b1300 b1302 b0961 b0960 b1050 b1113 b1498 b1867 b2347 b2409 b1596 b1276 b1478 b2078 b1710 b1990 b2439 b0810 b0809 b0654 b0653 b0652 b4428 b2075 b2076 b2077 b0783 b0784 b0785 b1420 b1127 b1743 b1519 b1197 b0662 b1376 b1477 b1535 b1598 b1706 b1792 b1917 b1918 b1919 b1956 b1976 b2301 b2410 b1045 b1046 b1330 b1520 b1595 | asr baeR glnH gltI helD mdtA putA putP puuB puuC puuE yccF yccS yceK ycfS ydeN yecD yfdC yfeR ynfM acnA adhP baeS btuE erfK eutL glnP glnQ gltJ gltK gltL hokB mdtB mdtC mdtD moaC moaD moaE mokB pepT spy tam treA ubiF uspF yddM ydeH ydgD ydiU yeaO yecC yecS dcyD yedQ mtfA yfcF yfeH ymdB ymdC ynaI yneE ynfL | acid shock-inducible periplasmic protein | DNA-binding response regulator in two-component regulatory system with BaeS | glutamine transporter subunit | glutamate and aspartate transporter subunit | DNA helicase IV | multidrug efflux system, subunit A | fused DNA-binding transcriptional regulator/proline dehydrogenase/pyrroline-5-carboxylate dehydrogenase | proline:sodium symporter | gamma-Glu-putrescine oxidase, FAD/NAD(P)-binding | gamma-Glu-gamma-aminobutyraldehyde dehydrogenase, NAD(P)H-dependent | GABA aminotransferase, PLP-dependent | conserved inner membrane protein | predicted inner membrane protein | predicted lipoprotein | conserved protein | predicted hydrolase | predicted DNA-binding transcriptional regulator | predicted transporter | aconitate hydratase 1 | ethanol-active dehydrogenase/acetaldehyde-active reductase | sensory histidine kinase in two-component regulatory system with BaeR | predicted glutathione peroxidase | conserved protein with NAD(P)-binding Rossmann-fold domain | predicted carboxysome structural protein with predicted role in ethanolamine utilization | toxic polypeptide, small | multidrug efflux system, subunit B | multidrug efflux system, subunit C | multidrug efflux system protein | molybdopterin biosynthesis, protein C | molybdopterin synthase, small subunit | molybdopterin synthase, large subunit | regulatory peptide | peptidase T | envelope stress induced periplasmic protein | trans-aconitate methyltransferase | periplasmic trehalase | 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol oxygenase | stress-induced protein, ATP-binding protein | predicted peptidase | predicted transporter subunit: ATP-binding component of ABC superfamily | predicted transporter subunit: membrane component of ABC superfamily | D-cysteine desulfhydrase, PLP-dependent | predicted diguanylate cyclase | predicted enzyme |
| 25 | 63 | 332 | -0.051 | 0.413 |  |  | 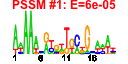 | 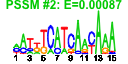 |  | b1982 b1693 b1704 b1709 b1249 b1191 b1275 b1538 b1844 b1920 b0810 b0809 b0652 b1635 b1842 b1174 b1740 b1708 b1603 b1658 b1854 b1608 b4432 b1642 b1059 b1682 b1034 b1087 b1056 b1057 b1177 b1178 b1179 b1180 b1282 b1251 b1248 b1328 b1381 b1461 b1431 b1510 b1539 b1607 b1703 b1794 b1833 b2152 b2154 b1045 b1046 b1382 b1448 b1520 b1793 b1612 b1611 b1613 b1494 b1609 b4529 b1447 b1495 | amn aroD aroH btuD cls cvrA cysB dcp exoX fliY glnP glnQ gltL gst holE minE nadE nlpC pntA purR pykA rstA slyA solA sufC ycdX yceF yceI yceJ ycgJ ycgK ycgL ycgM yciH yciI yciU ycjZ ydbH pptA ydcL ydeK ydfG ydgC ydiA yeaP yebS yeiB yeiG ymdB ymdC ynbE yncA yneE yoaF fumA fumC manA pqqL rstB ydbJ ydcZ yddB | AMP nucleosidase | 3-dehydroquinate dehydratase | 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase, tryptophan repressible | vitamin B12 transporter subunit : ATP-binding component of ABC superfamily | cardiolipin synthase 1 | predicted cation/proton antiporter | DNA-binding transcriptional dual regulator, O-acetyl-L-serine-binding | dipeptidyl carboxypeptidase II | DNA exonuclease X | cystine transporter subunit | glutamine transporter subunit | glutamate and aspartate transporter subunit | glutathionine S-transferase | DNA polymerase III, theta subunit | cell division topological specificity factor | NAD synthetase, NH3/glutamine-dependent | predicted lipoprotein | pyridine nucleotide transhydrogenase, alpha subunit | DNA-binding transcriptional repressor, hypoxanthine-binding | pyruvate kinase II | DNA-binding response regulator in two-component regulatory system with RstB | DNA-binding transcriptional activator | N-methyltryptophan oxidase, FAD-binding | component of SufBCD complex, ATP-binding component of ABC superfamily | predicted zinc-binding hydrolase | predicted protein | predicted cytochrome b561 | conserved protein | predicted isomerase/hydrolase | predicted enzyme | predicted DNA-binding transcriptional regulator | 4-oxalocrotonate tautomerase | L-allo-threonine dehydrogenase, NAD(P)-binding | conserved inner membrane protein associated with alginate biosynthesis | predicted diguanylate cyclase | conserved inner membrane protein | predicted esterase | predicted hydrolase | predicted acyltransferase with acyl-CoA N-acyltransferase domain | conserved outer membrane protein | fumarate hydratase (fumarase A), aerobic Class I | fumarate hydratase (fumarase C),aerobic Class II | mannose-6-phosphate isomerase | predicted peptidase | sensory histidine kinase in two-component regulatory system with RstA | predicted inner membrane protein | predicted porin protein |
| 46 | 66 | 341 | -0.014 | 0.478 |  |  | 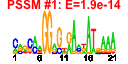 | 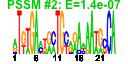 |  | b0863 b0862 b1710 b1661 b0882 b0881 b0880 b1952 b2441 b2439 b2000 b1064 b1619 b1250 b1827 b0870 b0878 b1065 b0781 b0783 b0784 b0785 b1068 b0957 b2418 b1127 b1066 b1324 b0711 b0712 b0713 b0869 b0965 b1067 b1100 b1104 b1108 b1614 b1664 b1792 b1837 b1917 b1918 b1919 b1971 b2001 b2301 b2419 b1449 b0605 b1709 b1275 b2133 b2440 b1920 b2442 b0782 b0760 b0714 b1981 b0710 b0946 b1962 b2438 b0502 b1448 | artI artQ btuE cfa clpA clpS cspD dsrB eutB eutL flu grxB hdhA kch kdgR ltaE macA mdtH moaA moaC moaD moaE yceM ompA pdxK pepT rimJ tpx ybgJ ybgK ybgL ybjT yccU yceH ycfH ycfL ycfP ydgA ydhQ yeaO yebW yecC yecS dcyD yedY yeeR yfcF yfeK yncB ahpC btuD cysB dld eutC fliY intZ moaB modF nei shiA ybgI ycbW yedJ eutK ylbG yncA | arginine transporter subunit | predicted glutathione peroxidase | cyclopropane fatty acyl phospholipid synthase (unsaturated-phospholipid methyltransferase) | ATPase and specificity subunit of ClpA-ClpP ATP-dependent serine protease, chaperone activity | regulatory protein for ClpA substrate specificity | cold shock protein homolog | predicted protein | ethanolamine ammonia-lyase, large subunit, heavy chain | predicted carboxysome structural protein with predicted role in ethanolamine utilization | CP4-44 prophage; antigen 43 (Ag43) phase-variable biofilm formation autotransporter | glutaredoxin 2 (Grx2) | 7-alpha-hydroxysteroid dehydrogenase, NAD-dependent | voltage-gated potassium channel | predicted DNA-binding transcriptional regulator | L-allo-threonine aldolase, PLP-dependent | macrolide transporter subunit, membrane fusion protein (MFP) component | predicted drug efflux system | molybdopterin biosynthesis protein A | molybdopterin biosynthesis, protein C | molybdopterin synthase, small subunit | molybdopterin synthase, large subunit | predicted oxidoreductase with NAD(P)-binding Rossmann-fold domain | outer membrane protein A (3a;II*;G;d) | pyridoxal-pyridoxamine kinase/hydroxymethylpyrimidine kinase | peptidase T | ribosomal-protein-S5-alanine N-acetyltransferase | lipid hydroperoxide peroxidase | predicted enzyme subunit | predicted lactam utilization protein | conserved protein with NAD(P)-binding Rossmann-fold domain | predicted CoA-binding protein with NAD(P)-binding Rossmann-fold domain | conserved protein | predicted metallodependent hydrolase | predicted transporter subunit: ATP-binding component of ABC superfamily | predicted transporter subunit: membrane component of ABC superfamily | D-cysteine desulfhydrase, PLP-dependent | predicted reductase | CP4-44 prophage; predicted membrane protein | predicted enzyme | predicted oxidoreductase, Zn-dependent and NAD(P)-binding | alkyl hydroperoxide reductase, C22 subunit | vitamin B12 transporter subunit : ATP-binding component of ABC superfamily | DNA-binding transcriptional dual regulator, O-acetyl-L-serine-binding | D-lactate dehydrogenase, FAD-binding, NADH independent | ethanolamine ammonia-lyase, small subunit (light chain) | cystine transporter subunit | CPZ-55 prophage; predicted integrase | molybdopterin biosynthesis protein B | fused molybdate transporter subunits of ABC superfamily: ATP-binding components | endonuclease VIII/ 5-formyluracil/5-hydroxymethyluracil DNA glycosylase | shikimate transporter | conserved metal-binding protein | predicted phosphohydrolase | predicted acyltransferase with acyl-CoA N-acyltransferase domain |
| 51 | 63 | 331 | -0.009 | 0.426 |  |  | 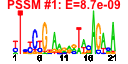 | 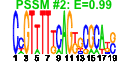 |  | b2296 b2901 b0733 b0734 b2369 b3963 b4232 b3894 b3893 b3892 b3177 b4443 b2905 b3176 b3413 b2947 b4349 b4350 b2747 b2746 b3617 b4129 b4242 b0009 b4351 b3208 b3961 b2743 b2908 b2829 b3782 b0414 b3017 b2744 b3616 b2745 b2907 b3831 b2906 b2532 b2909 b2946 b3646 b3755 b3964 b4234 b2900 b3042 b0114 b0115 b3209 b2748 b2828 b3967 b0237 b4235 b3041 b4373 b3591 b2827 b4374 b3011 b3207 | ackA bglA cydA cydB evgA fabR fbp fdoG fdoH fdoI folP gcvT glmM gntX gshB hsdM hsdR ispD ispF kbl lysU mgtA mog mrr mtgA oxyR pcm pepP ptsP ribD sufI surE tdh truD ubiH udp visC trmJ ygfB rsmE yicG yieP yijD yjgA yqfB yqiC aceE aceF elbB ftsB lgt murI pepD pmbA ribB rimI selA thyA yjjG yqhD yrbL | acetate kinase A and propionate kinase 2 | 6-phospho-beta-glucosidase A | cytochrome d terminal oxidase, subunit I | cytochrome d terminal oxidase, subunit II | DNA-binding response regulator in two-component regulatory system with EvgS | DNA-binding transcriptional repressor | fructose-1,6-bisphosphatase I | formate dehydrogenase-O, large subunit | formate dehydrogenase-O, Fe-S subunit | formate dehydrogenase-O, cytochrome b556 subunit | 7,8-dihydropteroate synthase | aminomethyltransferase, tetrahydrofolate-dependent, subunit (T protein) of glycine cleavage complex | phosphoglucosamine mutase | gluconate periplasmic binding protein with phosphoribosyltransferase domain, GNT I system | glutathione synthetase | DNA methylase M | endonuclease R | 4-diphosphocytidyl-2C-methyl-D-erythritol synthase | 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase | glycine C-acetyltransferase | lysine tRNA synthetase, inducible | magnesium transporter | predicted molybdochelatase | methylated adenine and cytosine restriction protein | biosynthetic peptidoglycan transglycosylase | DNA-binding transcriptional dual regulator | L-isoaspartate protein carboxylmethyltransferase type II | proline aminopeptidase P II | fused PTS enzyme: PEP-protein phosphotransferase (enzyme I)/GAF domain containing protein | fused diaminohydroxyphosphoribosylaminopyrimidine deaminase and 5-amino-6-(5-phosphoribosylamino) uracil reductase | repressor protein for FtsI | broad specificity 5'(3')-nucleotidase and polyphosphatase | threonine 3-dehydrogenase, NAD(P)-binding | pseudoruidine synthase | 2-octaprenyl-6-methoxyphenol hydroxylase, FAD/NAD(P)-binding | uridine phosphorylase | predicted oxidoreductase with FAD/NAD(P)-binding domain | predicted methyltransferase | predicted protein | conserved inner membrane protein | predicted transcriptional regulator | conserved protein | pyruvate dehydrogenase, decarboxylase component E1, thiamin-binding | pyruvate dehydrogenase, dihydrolipoyltransacetylase component E2 | isoprenoid biosynthesis protein with amidotransferase-like domain | cell division protein | phosphatidylglycerol-prolipoprotein diacylglyceryl transferase | glutamate racemase | aminoacyl-histidine dipeptidase (peptidase D) | predicted peptidase required for the maturation and secretion of the antibiotic peptide MccB17 | 3,4-dihydroxy-2-butanone-4-phosphate synthase | acetylase for 30S ribosomal subunit protein S18 | selenocysteine synthase | thymidylate synthetase | dUMP phosphatase | alcohol dehydrogenase, NAD(P)-dependent |
| 49 | 73 | 332 | -0.007 | 0.470 |  |  | 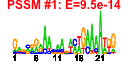 |  |  | b4088 b4086 b4085 b4084 b4017 b3690 b4478 b4320 b2873 b4266 b4285 b3710 b3422 b3583 b3582 b4303 b3061 b3062 b4194 b4198 b4192 b2870 b2871 b3060 b3086 b3382 b3586 b3587 b3597 b3598 b3689 b3696 b4482 b4274 b4308 b4309 b4337 b4486 b2874 b2941 b3013 b3047 b3050 b3051 b3393 b3394 b4230 b4087 b2934 b3693 b4479 b3691 b3900 b3391 b4267 b4268 b4090 b4565 b4304 b2866 b2984 b2985 b3518 b3656 b3657 b3711 b4083 b4311 b4310 b3152 b3392 b3395 b4485 | alsB alsC alsE alsK arpA cbrA dgoD fimH hyuA idnO mdtL rtcR sgbE sgbU sgcQ ttdA ttdB ulaB ulaF ulaG ygeW ygeX ttdR ygjQ yhfY yiaV yiaW yibH yibI yidR yidX yigE yjhR yjhS mdtM yjiV yqeA yqgD yqhG yqiH yqiJ yqiK hofO hofN ytfT alsA cmtB dgoK dgoR dgoT frvA hofQ idnD idnK rpiB sgcB sgcC xdhA yghR yghS yhjA yicI yicJ yidZ yjcS nanC yjhT yraR hofP hofM ytfR | D-allose transporter subunit | allulose-6-phosphate 3-epimerase | D-allose kinase | ankyrin repeat protein, function unknown | predicted oxidoreductase with FAD/NAD(P)-binding domain | galactonate dehydratase | minor component of type 1 fimbriae | D-stereospecific phenylhydantoinase | 5-keto-D-gluconate-5-reductase | multidrug efflux system protein | sigma 54-dependent transcriptional regulator of rtcBA expression | L-ribulose-5-phosphate 4-epimerase | predicted L-xylulose 5-phosphate 3-epimerase | KpLE2 phage-like element; predicted nucleoside triphosphatase | L-tartrate dehydratase, alpha subunit | L-tartrate dehydratase, beta subunit | L-ascorbate-specific enzyme IIB component of PTS | L-ribulose 5-phosphate 4-epimerase | predicted L-ascorbate 6-phosphate lactonase | conserved protein | 2,3-diaminopropionate ammonia-lyase | predicted DNA-binding transcriptional regulator | predicted thioredoxin-like protein | membrane fusion protein (MFP) component of efflux pump, signal anchor | conserved inner membrane protein | predicted protein | predicted inner membrane protein | predicted lipoproteinC | predicted amino acid kinase | predicted periplasmic pilin chaperone | conserved membrane protein | predicted fimbrial assembly protein | predicted sugar transporter subunit: membrane component of ABC superfamily | fused D-allose transporter subunits of ABC superfamily: ATP-binding components | predicted mannitol-specific enzyme IIA component of PTS | 2-oxo-3-deoxygalactonate kinase | D-galactonate transporter | predicted enzyme IIA component of PTS | predicted fimbrial transporter | L-idonate 5-dehydrogenase, NAD-binding | D-gluconate kinase, thermosensitive | ribose 5-phosphate isomerase B/allose 6-phosphate isomerase | predicted enzyme IIB component of PTS | KpLE2 phage-like element; predicted phosphotransferase enzyme IIC component | xanthine dehydrogenase, molybdenum binding subunit | predicted protein with nucleoside triphosphate hydrolase domain | predicted cytochrome C peroxidase | predicted alpha-glucosidase | predicted transporter | predicted alkyl sulfatase | N-acetylnuraminic acid outer membrane channel protein | predicted nucleoside-diphosphate-sugar epimerase | predicted pilus assembly protein | predicted sugar transporter subunit: ATP-binding component of ABC superfamily |
| 128 | 66 | 337 | 0.062 | 0.477 |  |  | 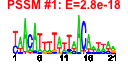 | 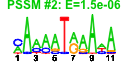 |  | b4069 b2310 b2309 b2307 b2306 b2308 b1297 b1298 b1422 b1958 b4067 b1415 b2155 b0598 b1189 b1190 b0596 b0595 b0593 b0583 b0594 b0586 b1805 b0221 b0584 b0592 b0585 b1102 b0805 b2976 b2977 b2241 b2243 b0720 b2537 b2149 b2148 b0348 b0349 b0350 b0352 b0351 b0346 b0679 b1020 b1014 b1015 b1301 b1300 b1302 b1299 b1421 b0597 b4512 b0581 b0804 b1019 b1018 b1057 b1113 b1423 b1959 b2341 b2342 b4068 b1596 | acs argT hisJ hisM hisP hisQ puuA puuD ydcI yedI actP aldA cirA cstA dadA dadX entA entB entC entD entE entF fadD fadE fepA fepB fes fhuE fiu glcB glcG glpA glpC gltA hcaR mglA mglC mhpB mhpC mhpD mhpE mhpF mhpR nagE phoH putA putP puuB puuC puuE puuR trg ybdB ybdD ybdK ybiX ycdB ycdO yceJ ycfS ydcJ yedA fadJ fadI yjcH ynfM | acetyl-CoA synthetase | lysine/arginine/ornithine transporter subunit | histidine/lysine/arginine/ornithine transporter subunit | gamma-Glu-putrescine synthase | gamma-Glu-GABA hydrolase | predicted DNA-binding transcriptional regulator | conserved inner membrane protein | acetate transporter | aldehyde dehydrogenase A, NAD-linked | ferric iron-catecholate outer membrane transporter | carbon starvation protein | D-amino acid dehydrogenase | alanine racemase 2, PLP-binding | 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase | isochorismatase | isochorismate synthase 1 | phosphopantetheinyltransferase component of enterobactin synthase multienzyme complex | 2,3-dihydroxybenzoate-AMP ligase component of enterobactin synthase multienzyme complex | enterobactin synthase multienzyme complex component, ATP-dependent | acyl-CoA synthetase (long-chain-fatty-acid--CoA ligase) | acyl coenzyme A dehydrogenase | iron-enterobactin outer membrane transporter | iron-enterobactin transporter subunit | enterobactin/ferric enterobactin esterase | ferric-rhodotorulic acid outer membrane transporter | predicted iron outer membrane transporter | malate synthase G | conserved protein | sn-glycerol-3-phosphate dehydrogenase (anaerobic), large subunit, FAD/NAD(P)-binding | sn-glycerol-3-phosphate dehydrogenase (anaerobic), small subunit | citrate synthase | DNA-binding transcriptional activator of 3-phenylpropionic acid catabolism | fused methyl-galactoside transporter subunits of ABC superfamily: ATP-binding components | methyl-galactoside transporter subunit | 2,3-dihydroxyphenylpropionate 1,2-dioxygenase | 2-hydroxy-6-ketonona-2,4-dienedioic acid hydrolase | 2-keto-4-pentenoate hydratase | 4-hyroxy-2-oxovalerate/4-hydroxy-2-oxopentanoic acid aldolase, class I | acetaldehyde-CoA dehydrogenase II, NAD-binding | DNA-binding transcriptional activator, 3HPP-binding | fused N-acetyl glucosamine specific PTS enzyme: IIC, IIB , and IIA components | conserved protein with nucleoside triphosphate hydrolase domain | fused DNA-binding transcriptional regulator/proline dehydrogenase/pyrroline-5-carboxylate dehydrogenase | proline:sodium symporter | gamma-Glu-putrescine oxidase, FAD/NAD(P)-binding | gamma-Glu-gamma-aminobutyraldehyde dehydrogenase, NAD(P)H-dependent | GABA aminotransferase, PLP-dependent | DNA-binding transcriptional repressor | methyl-accepting chemotaxis protein III, ribose and galactose sensor receptor | gamma-glutamyl:cysteine ligase | predicted cytochrome b561 | predicted inner membrane protein | fused enoyl-CoA hydratase and epimerase and isomerase/3-hydroxyacyl-CoA dehydrogenase | beta-ketoacyl-CoA thiolase, anaerobic, subunit | conserved inner membrane protein involved in acetate transport | predicted transporter |
| 106 | 65 | 340 | 0.069 | 0.491 |  |  | 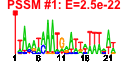 | 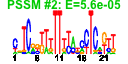 |  | b3571 b4103 b4031 b3504 b4050 b4066 b3265 b3266 b3138 b3140 b2918 b3801 b3077 b3264 b3899 b3683 b2484 b4076 b4102 b3379 b2826 b2825 b2823 b3904 b3902 b3905 b3907 b3422 b3567 b3568 b2505 b2577 b2578 b2778 b2824 b2920 b2888 b3078 b3079 b3104 b3262 b3369 b3376 b3377 b3378 b3380 b3381 b3382 b3383 b3586 b3587 b3597 b3598 b3874 b4186 b4182 b4184 b4185 b4257 b4340 b4341 b2917 b2775 b3393 b4002 | malS xylE yhiS pspG yjcF acrE acrF agaB agaD argK aslA ebgC envR frvB glvC hyfD nrfG phnF php ppdA ppdB ppdC rhaB rhaD rhaS rhaT rtcR xylG xylH yfgH yfiE eamB ygcG ygdB scpC ygfU ygjI ygjJ yhaI yhdJ yhfL yhfS yhfT yhfU yhfW yhfX yhfY yhfZ yiaV yiaW yibH yibI yihN yjfC yjfJ yjfL yjfM yjgN yjiR yjiS scpA yqcE hofO zraP | alpha-amylase | D-xylose transporter | phage shock protein G | conserved protein | cytoplasmic membrane lipoprotein | multidrug efflux system protein | N-acetylgalactosamine-specific enzyme IIB component of PTS | N-acetylgalactosamine-specific enzyme IID component of PTS | membrane ATPase/protein kinase | acrylsulfatase-like enzyme | cryptic beta-D-galactosidase, beta subunit | DNA-binding transcriptional regulator | fused predicted PTS enzymes: IIB component/IIC component | hydrogenase 4, membrane subunit | heme lyase (NrfEFG) for insertion of heme into c552, subunit NrfG | predicted DNA-binding transcriptional regulator of phosphonate uptake and biodegradation | predicted hydrolase | predicted protein | rhamnulokinase | rhamnulose-1-phosphate aldolase | DNA-binding transcriptional activator, L-rhamnose-binding | L-rhamnose:proton symporter | sigma 54-dependent transcriptional regulator of rtcBA expression | fused D-xylose transporter subunits of ABC superfamily: ATP-binding components | D-xylose transporter subunit | predicted outer membrane lipoprotein | predicted DNA-binding transcriptional regulator | neutral amino-acid efflux system | propionyl-CoA:succinate-CoA transferase | predicted transporter | predicted inner membrane protein | predicted methyltransferase | conserved secreted peptide | predicted mutase | predicted amino acid racemase | membrane fusion protein (MFP) component of efflux pump, signal anchor | conserved inner membrane protein | predicted synthetase/amidase | predicted transcriptional regulator effector protein | fused predicted DNA-binding transcriptional regulator/predicted aminotransferase | methylmalonyl-CoA mutase | conserved membrane protein | Zn-binding periplasmic protein |
| 30 | 70 | 335 | 0.091 | 0.429 |  |  | 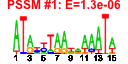 | 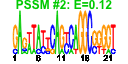 |  | b3138 b2934 b0140 b4313 b2374 b3875 b2854 b0254 b2648 b2734 b4300 b0135 b0136 b0141 b0218 b0358 b0364 b0375 b4579 b0558 b0603 b0629 b0705 b4514 b1347 b4526 b2273 b2274 b2275 b2345 b2503 b2625 b2626 b2850 b2851 b2852 b2853 b2855 b2856 b2956 b2968 b2984 b2985 b2998 b3215 b3216 b3262 b3270 b3615 b4083 b4183 b4333 b4365 b0370 b2548 b2846 b2849 b3046 b3048 b3142 b3143 b3392 b3443 b2933 b3897 b3269 b3271 b3442 b4332 b4210 | agaB cmtB ecpD fimE frc ompL pbl perR pinH pphB sgcR yadC yadK yadN yafU yaiO yaiS yaiV yaiX ybcV ybdO ybeF ybfL ybfQ ydaC ydaE yfbN yfbO yfbP yfdF yfgF yfjI yfjJ ygeF ygeG ygeH ygeI ygeK yggM yghD yghR yghS yghW yhcA yhcD yhdJ yhdY yibD yjcS yjfK yjiK yjjQ yphF yqeH yqeK yqiG yqiI yraH yraI hofP yrhA cmtA frvR yhdX yhdZ yhhZ yjiJ ytfF | N-acetylgalactosamine-specific enzyme IIB component of PTS | predicted mannitol-specific enzyme IIA component of PTS | predicted periplasmic pilin chaperone | tyrosine recombinase/inversion of on/off regulator of fimA | formyl-CoA transferase, NAD(P)-binding | predicted outer membrane porin L | CP4-6 prophage; predicted DNA-binding transcriptional regulator | serine/threonine-specific protein phosphatase 2 | KpLE2 phage-like element; predicted DNA-binding transcriptional regulator | predicted fimbrial-like adhesin protein | predicted protein | conserved protein | predicted DNA-binding transcriptional regulator | DLP12 prophage; predicted protein | predicted transposase | Rac prophage; predicted protein | Rac prophage; conserved protein | predicted inner membrane protein | CP4-57 prophage; predicted protein | predicted chaperone | predictedtranscriptional regulator | predicted secretion pathway M-type protein, membrane anchored | predicted protein with nucleoside triphosphate hydrolase domain | predicted periplasmic chaperone protein | predicted outer membrane protein | predicted methyltransferase | predicted amino-acid transporter subunit | predicted glycosyl transferase | predicted alkyl sulfatase | predicted sugar transporter subunit: periplasmic-binding component of ABC superfamily | conserved protein with bipartite regulator domain | predicted fused mannitol-specific PTS enzymes: IIB component/IIC component | predicted regulator |
| 136 | 83 | 330 | 0.099 | 0.480 |  |  | 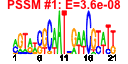 | 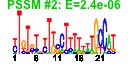 |  | b2329 b2328 b2330 b2174 b2327 b2325 b2326 b2697 b2480 b0432 b0431 b0430 b0429 b0428 b2478 b3963 b4314 b4315 b2303 b1905 b1779 b2094 b2093 b2092 b2095 b2904 b2479 b2905 b2508 b0104 b1237 b0440 b0116 b1817 b1818 b0963 b1176 b1175 b0827 b2518 b2477 b0553 b2288 b2287 b2286 b2285 b2284 b2283 b2282 b2281 b2280 b2279 b2278 b2277 b2393 b0565 b2564 b1278 b0931 b2415 b1277 b0721 b0722 b0893 b0921 b1656 b2175 b3962 b0726 b0727 b0729 b0411 b1914 b2346 b0410 b0877 b0970 b2007 b2293 b2294 b2331 b2595 b3964 | aroC mepA prmB yeiU yfcA yfcL yfcM alaS bcp cyoA cyoB cyoC cyoD cyoE dapA fabR fimA fimI folX ftnA gapA gatA gatB gatC gatZ gcvH gcvR gcvT guaB guaC hns hupB lpd manX manY mgsA minC minD moeA ndk nlpB nmpC nuoA nuoB nuoC nuoE nuoF nuoG nuoH nuoI nuoJ nuoK nuoL nuoM nupC ompT pdxJ pgpB pncB ptsH ribA sdhC sdhD serS smtA sodB spr sthA sucA sucB sucD tsx uvrY vacJ yajD ybjX yccA yeeX yfbT yfbU yfcN yfiO yijD | chorismate synthase | murein DD-endopeptidase | N5-glutamine methyltransferase | undecaprenyl pyrophosphate phosphatase | conserved inner membrane protein | predicted protein | conserved protein | alanyl-tRNA synthetase | thiol peroxidase, thioredoxin-dependent | cytochrome o ubiquinol oxidase subunit II | cytochrome o ubiquinol oxidase subunit I | cytochrome o ubiquinol oxidase subunit III | cytochrome o ubiquinol oxidase subunit IV | protoheme IX farnesyltransferase | dihydrodipicolinate synthase | DNA-binding transcriptional repressor | major type 1 subunit fimbrin (pilin) | fimbrial protein involved in type 1 pilus biosynthesis | D-erythro-7,8-dihydroneopterin triphosphate 2'-epimerase and dihydroneopterin aldolase | ferritin iron storage protein (cytoplasmic) | glyceraldehyde-3-phosphate dehydrogenase A | galactitol-specific enzyme IIA component of PTS | galactitol-specific enzyme IIB component of PTS | galactitol-specific enzyme IIC component of PTS | D-tagatose 1,6-bisphosphate aldolase 2, subunit | glycine cleavage complex lipoylprotein | DNA-binding transcriptional repressor, regulatory protein accessory to GcvA | aminomethyltransferase, tetrahydrofolate-dependent, subunit (T protein) of glycine cleavage complex | IMP dehydrogenase | GMP reductase | global DNA-binding transcriptional dual regulator H-NS | HU, DNA-binding transcriptional regulator, beta subunit | lipoamide dehydrogenase, E3 component is part of three enzyme complexes | fused mannose-specific PTS enzymes: IIA component/IIB component | mannose-specific enzyme IIC component of PTS | methylglyoxal synthase | cell division inhibitor | membrane ATPase of the MinC-MinD-MinE system | molybdopterin biosynthesis protein | multifunctional nucleoside diphosphate kinase and apyrimidinic endonuclease and 3'-phosphodiesterase | lipoprotein | NADH:ubiquinone oxidoreductase, membrane subunit A | NADH:ubiquinone oxidoreductase, chain B | NADH:ubiquinone oxidoreductase, chain C,D | NADH:ubiquinone oxidoreductase, chain E | NADH:ubiquinone oxidoreductase, chain F | NADH:ubiquinone oxidoreductase, chain G | NADH:ubiquinone oxidoreductase, membrane subunit H | NADH:ubiquinone oxidoreductase, chain I | NADH:ubiquinone oxidoreductase, membrane subunit J | NADH:ubiquinone oxidoreductase, membrane subunit K | NADH:ubiquinone oxidoreductase, membrane subunit L | NADH:ubiquinone oxidoreductase, membrane subunit M | nucleoside (except guanosine) transporter | DLP12 prophage; outer membrane protease VII (outer membrane protein 3b) | pyridoxine 5'-phosphate synthase | phosphatidylglycerophosphatase B | nicotinate phosphoribosyltransferase | phosphohistidinoprotein-hexose phosphotransferase component of PTS system (Hpr) | GTP cyclohydrolase II | succinate dehydrogenase, membrane subunit, binds cytochrome b556 | seryl-tRNA synthetase, also charges selenocysteinyl-tRNA with serine | predicted S-adenosyl-L-methionine-dependent methyltransferase | superoxide dismutase, Fe | predicted peptidase, outer membrane lipoprotein | pyridine nucleotide transhydrogenase, soluble | 2-oxoglutarate decarboxylase, thiamin-requiring | dihydrolipoyltranssuccinase | succinyl-CoA synthetase, NAD(P)-binding, alpha subunit | nucleoside channel, receptor of phage T6 and colicin K | DNA-binding response regulator in two-component regulatory system with BarA | predicted lipoprotein | inner membrane protein | predicted hydrolase or phosphatase |
| 149 | 69 | 336 | 0.210 | 0.443 |  |  | 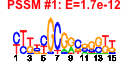 | 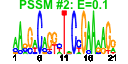 |  | b4053 b3918 b4382 b4383 b4381 b4384 b3066 b2798 b3891 b3894 b3893 b3892 b4291 b4290 b4287 b0356 b0355 b0357 b3423 b4077 b3500 b2947 b2708 b3401 b3400 b3932 b0182 b0459 b2265 b0009 b4233 b3250 b3635 b3163 b3183 b3019 b3493 b3863 b3498 b2679 b0405 b3822 b3778 b3623 b3789 b0183 b3461 b2944 b3017 b3706 b3831 b2951 b2953 b2954 b2955 b3084 b3184 b3343 b3352 b3492 b3497 b3499 b3529 b3539 b3552 b4176 b4234 b2690 b3399 | alr cdh deoA deoB deoC deoD dnaG xni fdhE fdoG fdoH fdoI fecA fecB fecE frmA frmB frmR glpR gltP gor gshB gutQ hslO hslR hslV lpxB maa menF mog mpl mreC mutM nlpI obgE parC pitA polA prlC proX queA recQ rep waaU rffH rnhB rpoH yggI sufI mnmE udp yggS yggU rdgB yggW rlmG yhbE yheL yheS yhiN yhiQ yhiR yhjK yhjV yiaD yjeT yjgA yqaB yrfG | alanine racemase 1, PLP-binding, biosynthetic | CDP-diacylglycerol phosphotidylhydrolase | thymidine phosphorylase | phosphopentomutase | 2-deoxyribose-5-phosphate aldolase, NAD(P)-linked | purine-nucleoside phosphorylase | DNA primase | exonuclease IX (5'-3' exonuclease) | formate dehydrogenase formation protein | formate dehydrogenase-O, large subunit | formate dehydrogenase-O, Fe-S subunit | formate dehydrogenase-O, cytochrome b556 subunit | KpLE2 phage-like element; ferric citrate outer membrane transporter | KpLE2 phage-like element; iron-dicitrate transporter subunit | alcohol dehydrogenase class III/glutathione-dependent formaldehyde dehydrogenase | predicted esterase | regulator protein that represses frmRAB operon | DNA-binding transcriptional repressor | glutamate/aspartate:proton symporter | glutathione oxidoreductase | glutathione synthetase | predicted phosphosugar-binding protein | heat shock protein Hsp33 | ribosome-associated heat shock protein Hsp15 | peptidase component of the HslUV protease | tetraacyldisaccharide-1-P synthase | maltose O-acetyltransferase | isochorismate synthase 2 | predicted molybdochelatase | UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl- meso-diaminopimelate ligase | cell wall structural complex MreBCD transmembrane component MreC | formamidopyrimidine/5-formyluracil/ 5-hydroxymethyluracil DNA glycosylase | conserved protein | GTPase involved in cell partioning and DNA repair | DNA topoisomerase IV, subunit A | phosphate transporter, low-affinity | fused DNA polymerase I 5'->3' exonuclease/3'->5' polymerase/3'->5' exonuclease | oligopeptidase A | glycine betaine transporter subunit | S-adenosylmethionine:tRNA ribosyltransferase-isomerase | ATP-dependent DNA helicase | DNA helicase and single-stranded DNA-dependent ATPase | lipopolysaccharide core biosynthesis | glucose-1-phosphate thymidylyltransferase | ribonuclease HII, degrades RNA of DNA-RNA hybrids | RNA polymerase, sigma 32 (sigma H) factor | repressor protein for FtsI | GTPase | uridine phosphorylase | predicted enzyme | dITP/XTP pyrophosphatase | predicted oxidoreductase | predicted methyltransferase small domain | conserved inner membrane protein | predicted intracellular sulfur oxidation protein | fused predicted transporter subunits of ABC superfamily: ATP-binding components | predicted oxidoreductase with FAD/NAD(P)-binding domain | predicted SAM-dependent methyltransferase | predicted DNA (exogenous) processing protein | predicted diguanylate cyclase | predicted transporter | predicted outer membrane lipoprotein | predicted hydrolase |
| 26 | 53 | 336 | 0.222 | 0.474 |  |  | 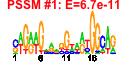 | 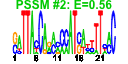 |  | b1276 b0118 b0908 b0928 b0839 b2133 b2153 b0720 b0653 b2551 b0755 b0828 b1136 b1209 b2156 b3236 b0929 b1602 b0386 b0945 b2218 b0907 b1981 b0002 b0003 b0004 b0387 b0801 b0874 b0946 b1033 b1287 b1729 b1778 b1777 b2012 b2013 b2495 b0829 b0830 b0831 b0838 b1143 b1982 b1603 b1608 b4432 b1056 b1251 b1461 b1607 b2152 b2154 | acnA acnB aroA aspC dacC dld folE gltA gltK glyA gpmA iaaA icd lolB lysP mdh ompF pntB proC pyrD rcsC serC shiA thrA thrB thrC yaiI ybiC ybjE ycbW ghrA yciW ydjN msrB yeaC yeeD yeeE yfgD gsiA gsiB gsiC yliJ ymfI amn pntA rstA yceI yciI pptA ydgC yeiB yeiG | aconitate hydratase 1 | bifunctional aconitate hydratase 2/2-methylisocitrate dehydratase | 5-enolpyruvylshikimate-3-phosphate synthetase | aspartate aminotransferase, PLP-dependent | D-alanyl-D-alanine carboxypeptidase (penicillin-binding protein 6a) | D-lactate dehydrogenase, FAD-binding, NADH independent | GTP cyclohydrolase I | citrate synthase | glutamate and aspartate transporter subunit | serine hydroxymethyltransferase | phosphoglyceromutase 1 | L-asparaginase | e14 prophage; isocitrate dehydrogenase, specific for NADP+ | chaperone for lipoproteins | lysine transporter | malate dehydrogenase, NAD(P)-binding | outer membrane porin 1a (Ia;b;F) | pyridine nucleotide transhydrogenase, beta subunit | pyrroline-5-carboxylate reductase, NAD(P)-binding | dihydro-orotate oxidase, FMN-linked | hybrid sensory kinase in two-component regulatory system with RcsB and YojN | 3-phosphoserine/phosphohydroxythreonine aminotransferase | shikimate transporter | fused aspartokinase I and homoserine dehydrogenase I | homoserine kinase | threonine synthase | conserved protein | predicted dehydrogenase | predicted transporter | predicted protein | 2-ketoacid reductase | predicted oxidoreductase | methionine sulfoxide reductase B | predicted inner membrane protein | fused predicted peptide transport subunits of ABC superfamily: ATP-binding components | predicted peptide transporter subunit: periplasmic-binding component of ABC superfamily | predicted peptide transporter subunit: membrane component of ABC superfamily | predicted glutathione S-transferase | e14 prophage; predicted protein | AMP nucleosidase | pyridine nucleotide transhydrogenase, alpha subunit | DNA-binding response regulator in two-component regulatory system with RstB | predicted enzyme | 4-oxalocrotonate tautomerase | conserved inner membrane protein associated with alginate biosynthesis | conserved inner membrane protein | predicted esterase |
| 141 | 73 | 333 | 0.270 | 0.480 |  |  | 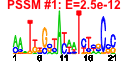 | 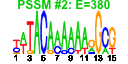 |  | b3256 b0474 b0469 b2329 b2317 b1850 b1851 b2685 b2566 b2323 b0684 b2321 b0529 b1210 b0640 b0884 b1718 b2747 b2746 b2569 b2568 b0642 b0630 b2890 b2328 b2114 b0634 b2113 b0639 b2234 b2235 b0416 b2743 b2320 b1714 b2330 b2780 b2565 b0472 b3052 b0415 b0633 b0641 b3386 b2914 b1716 b1717 b0120 b0121 b2744 b1719 b2935 b2318 b2745 b0888 b2319 b0434 b0471 b0413 b0636 b0637 b0853 b1213 b2236 b2327 b2299 b2300 b2326 b2350 b2946 b2611 b2948 b3002 | accC adk apt aroC dedA eda edd emrA era fabB fldA flk folD hemA holA infA infC ispD ispF lepA lepB leuS lipB lysS mepA metG mrdB mrp nadD nrdA nrdB nusB pcm pdxB pheS prmB pyrG recO recR rfaE ribE rlpA rlpB rpe rpiA rplT rpmI speD speE surE thrS tktA truA truD trxB usg yajG ybaB nrdR ybeA ybeB ybjN ychQ yfaE yfcA yfcD yfcE yfcM yfdG rsmE ypjD yqgE yqhA | acetyl-CoA carboxylase, biotin carboxylase subunit | adenylate kinase | adenine phosphoribosyltransferase | chorismate synthase | conserved inner membrane protein | multifunctional 2-keto-3-deoxygluconate 6-phosphate aldolase and 2-keto-4-hydroxyglutarate aldolase and oxaloacetate decarboxylase | 6-phosphogluconate dehydratase | multidrug efflux system | membrane-associated, 16S rRNA-binding GTPase | 3-oxoacyl-[acyl-carrier-protein] synthase I | flavodoxin 1 | predicted flagella assembly protein | bifunctional 5,10-methylene-tetrahydrofolate dehydrogenase/ 5,10-methylene-tetrahydrofolate cyclohydrolase | glutamyl tRNA reductase | DNA polymerase III, delta subunit | translation initiation factor IF-1 | protein chain initiation factor IF-3 | 4-diphosphocytidyl-2C-methyl-D-erythritol synthase | 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase | GTP-binding membrane protein | leader peptidase (signal peptidase I) | leucyl-tRNA synthetase | lipoyl-protein ligase | lysine tRNA synthetase, constitutive | murein DD-endopeptidase | methionyl-tRNA synthetase | cell wall shape-determining protein | antiporter inner membrane protein | nicotinic acid mononucleotide adenylyltransferase, NAD(P)-dependent | ribonucleoside diphosphate reductase 1, alpha subunit | ribonucleoside diphosphate reductase 1, beta subunit, ferritin-like protein | transcription antitermination protein | L-isoaspartate protein carboxylmethyltransferase type II | erythronate-4-phosphate dehydrogenase | phenylalanine tRNA synthetase, alpha subunit | N5-glutamine methyltransferase | CTP synthetase | gap repair protein | fused heptose 7-phosphate kinase/heptose 1-phosphate adenyltransferase | riboflavin synthase beta chain | minor lipoprotein | D-ribulose-5-phosphate 3-epimerase | ribose 5-phosphate isomerase, constitutive | 50S ribosomal subunit protein L20 | 50S ribosomal subunit protein L35 | S-adenosylmethionine decarboxylase | spermidine synthase (putrescine aminopropyltransferase) | broad specificity 5'(3')-nucleotidase and polyphosphatase | threonyl-tRNA synthetase | transketolase 1, thiamin-binding | pseudouridylate synthase I | pseudoruidine synthase | thioredoxin reductase, FAD/NAD(P)-binding | predicted semialdehyde dehydrogenase | predicted lipoprotein | conserved protein | predicted protein | predicted oxidoreductase | predicted transcriptional regulator | predicted 2Fe-2S cluster-containing protein | predicted NUDIX hydrolase | predicted phosphatase | CPS-53 (KpLE1) prophage; bactoprenol-linked glucose translocase (flippase) | predicted inner membrane protein |
| 70 | 64 | 335 | 0.282 | 0.482 |  |  | 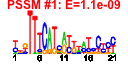 | 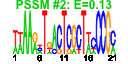 |  | b3241 b3265 b3266 b3234 b4079 b3447 b3580 b4074 b4075 b4076 b4097 b4304 b4305 b3453 b3567 b3568 b3448 b3579 b4080 b4081 b4082 b4186 b4231 b4338 b4002 b3588 b4088 b4086 b4084 b3722 b3720 b2979 b2713 b4266 b4264 b3647 b4005 b3421 b3581 b4302 b4301 b4303 b4300 b3452 b3450 b3451 b3449 b3566 b4465 b2975 b3106 b3268 b3270 b3524 b3684 b3718 b3719 b4190 b4334 b4335 b4506 b3394 b4230 b4003 | aaeA acrE acrF degQ fdhF ggt lyx nrfE nrfF nrfG phnK sgcC sgcX ugpB xylG xylH yhhA yiaO mdtP mdtO mdtN yjfC yjfF zraP aldB alsB alsC alsK bglF bglH glcD hydN idnO idnR ligB purD rtcB sgbH sgcA sgcE sgcQ sgcR ugpA ugpC ugpE ugpQ xylF yggP glcA yhaK yhdW yhdY yhjG yidP yieK yieL yjfP yjiL yjiM ykgO hofN ytfT zraS | p-hydroxybenzoic acid efflux system component | cytoplasmic membrane lipoprotein | multidrug efflux system protein | serine endoprotease, periplasmic | formate dehydrogenase-H, selenopolypeptide subunit | gamma-glutamyltranspeptidase | L-xylulose kinase | heme lyase (NrfEFG) for insertion of heme into c552, subunit NrfE | heme lyase (NrfEFG) for insertion of heme into c552, subunit NrfF | heme lyase (NrfEFG) for insertion of heme into c552, subunit NrfG | carbon-phosphorus lyase complex subunit | KpLE2 phage-like element; predicted phosphotransferase enzyme IIC component | KpLE2 phage-like element; predicted endoglucanase with Zn-dependent exopeptidase domain | glycerol-3-phosphate transporter subunit | fused D-xylose transporter subunits of ABC superfamily: ATP-binding components | D-xylose transporter subunit | conserved protein | predicted transporter | predicted outer membrane factor of efflux pump | predicted multidrug efflux system component | predicted membrane fusion protein of efflux pump | predicted synthetase/amidase | predicted sugar transporter subunit: membrane component of ABC superfamily | Zn-binding periplasmic protein | aldehyde dehydrogenase B | D-allose transporter subunit | D-allose kinase | fused beta-glucoside-specific PTS enzymes: IIA component/IIB component/IIC component | carbohydrate-specific outer membrane porin, cryptic | glycolate oxidase subunit, FAD-linked | formate dehydrogenase-H, [4Fe-4S] ferredoxin subunit | 5-keto-D-gluconate-5-reductase | DNA-binding transcriptional repressor, 5-gluconate-binding | DNA ligase, NAD(+)-dependent | phosphoribosylglycinamide synthetase phosphoribosylamine-glycine ligase | 3-keto-L-gulonate 6-phosphate decarboxylase | KpLE2 phage-like element; predicted phosphotransferase enzyme IIA component | KpLE2 phage-like element; predicted epimerase | KpLE2 phage-like element; predicted nucleoside triphosphatase | KpLE2 phage-like element; predicted DNA-binding transcriptional regulator | glycerophosphodiester phosphodiesterase, cytosolic | predicted dehydrogenase | glycolate transporter | predicted pirin-related protein | predicted amino-acid transporter subunit | predicted outer membrane biogenesis protein | predicted DNA-binding transcriptional regulator | predicted 6-phosphogluconolactonase | predicted xylanase | predicted hydrolase | predicted ATPase, activator of (R)-hydroxyglutaryl-CoA dehydratase | predicted 2-hydroxyglutaryl-CoA dehydratase | rpmJ (L36) paralog | predicted fimbrial assembly protein | predicted sugar transporter subunit: membrane component of ABC superfamily | sensory histidine kinase in two-component regulatory system with ZraR |
| 62 | 69 | 334 | 0.287 | 0.467 |  |  | 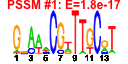 | 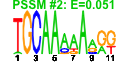 |  | b1094 b0032 b0033 b1216 b0337 b1823 b2313 b0632 b0031 b2553 b3212 b2507 b2552 b1718 b0198 b2942 b0199 b0197 b1245 b1246 b1101 b0523 b2312 b4006 b0522 b2499 b2500 b1849 b1232 b1062 b1716 b1717 b1234 b0739 b2498 b2497 b2346 b2509 b0736 b0772 b1832 b2290 b3714 b0908 b0928 b2414 b2153 b2551 b1136 b0889 b2156 b0929 b1243 b1244 b1247 b0741 b0945 b0907 b0002 b0003 b0004 b0740 b2580 b0742 b0874 b0926 b1729 b1143 b1824 | acpP carA carB chaA codA cspC cvpA dacA dapB glnB gltB guaA hmp infC metI metK metN metQ oppC oppD ptsG purE purF purH purK purM purN purT purU pyrC rplT rpmI rssA tolA upp uraA vacJ xseA ybgC ybhC yebR yfbQ yieG aroA aspC cysK folE glyA icd lrp lysP ompF oppA oppB oppF pal pyrD serC thrA thrB thrC tolB ung ybgF ybjE ycbK ydjN ymfI yobF | acyl carrier protein (ACP) | carbamoyl phosphate synthetase small subunit, glutamine amidotransferase | carbamoyl-phosphate synthase large subunit | calcium/sodium:proton antiporter | cytosine deaminase | stress protein, member of the CspA-family | membrane protein required for colicin V production | D-alanyl-D-alanine carboxypeptidase (penicillin-binding protein 5) | dihydrodipicolinate reductase | regulatory protein P-II for glutamine synthetase | glutamate synthase, large subunit | GMP synthetase (glutamine aminotransferase) | fused nitric oxide dioxygenase/dihydropteridine reductase 2 | protein chain initiation factor IF-3 | DL-methionine transporter subunit | methionine adenosyltransferase 1 | oligopeptide transporter subunit | fused glucose-specific PTS enzymes: IIB component/IIC component | N5-carboxyaminoimidazole ribonucleotide mutase | amidophosphoribosyltransferase | fused IMP cyclohydrolase/phosphoribosylaminoimidazolecarboxamide formyltransferase | N5-carboxyaminoimidazole ribonucleotide synthase | phosphoribosylaminoimidazole synthetase | phosphoribosylglycinamide formyltransferase 1 | phosphoribosylglycinamide formyltransferase 2 | formyltetrahydrofolate hydrolase | dihydro-orotase | 50S ribosomal subunit protein L20 | 50S ribosomal subunit protein L35 | conserved protein | membrane anchored protein in TolA-TolQ-TolR complex | uracil phosphoribosyltransferase | uracil transporter | predicted lipoprotein | exonuclease VII, large subunit | predicted acyl-CoA thioesterase | predicted pectinesterase | predicted aminotransferase | predicted inner membrane protein | 5-enolpyruvylshikimate-3-phosphate synthetase | aspartate aminotransferase, PLP-dependent | cysteine synthase A, O-acetylserine sulfhydrolase A subunit | GTP cyclohydrolase I | serine hydroxymethyltransferase | e14 prophage; isocitrate dehydrogenase, specific for NADP+ | DNA-binding transcriptional dual regulator, leucine-binding | lysine transporter | outer membrane porin 1a (Ia;b;F) | peptidoglycan-associated outer membrane lipoprotein | dihydro-orotate oxidase, FMN-linked | 3-phosphoserine/phosphohydroxythreonine aminotransferase | fused aspartokinase I and homoserine dehydrogenase I | homoserine kinase | threonine synthase | periplasmic protein | uracil-DNA-glycosylase | predicted protein | predicted transporter | e14 prophage; predicted protein |
| 121 | 80 | 320 | 0.307 | 0.478 |  |  | 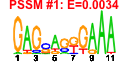 |  |  | b0432 b0431 b0430 b0429 b0428 b3528 b2094 b2093 b2092 b2095 b0116 b1817 b1818 b1819 b0963 b2518 b0553 b2288 b2287 b2286 b2285 b2284 b2283 b2282 b2281 b2280 b2279 b2278 b2277 b2393 b2259 b2415 b2797 b2796 b0721 b0722 b1656 b3962 b0726 b0727 b0729 b2935 b0411 b2007 b0118 b2209 b4152 b2091 b2096 b2388 b3927 b3926 b2614 b2463 b3417 b3416 b3236 b2276 b1245 b1246 b3403 b2523 b1308 b2416 b3749 b3751 b3750 b3748 b3752 b0651 b0610 b0723 b0724 b4388 b2521 b0728 b4240 b4239 b3646 b4352 | cyoA cyoB cyoC cyoD cyoE dctA gatA gatB gatC gatZ lpd manX manY manZ mgsA ndk nmpC nuoA nuoB nuoC nuoE nuoF nuoG nuoH nuoI nuoJ nuoK nuoL nuoM nupC pmrD ptsH sdaB sdaC sdhC sdhD sodB sthA sucA sucB sucD tktA tsx yeeX acnB eco frdC gatD gatY glk glpF glpK grpE maeB malP malQ mdh nuoN oppC oppD pck pepB pspE ptsI rbsA rbsB rbsC rbsD rbsK rihA rnk sdhA sdhB serB sseA sucC treB treC yicG yjiA | cytochrome o ubiquinol oxidase subunit II | cytochrome o ubiquinol oxidase subunit I | cytochrome o ubiquinol oxidase subunit III | cytochrome o ubiquinol oxidase subunit IV | protoheme IX farnesyltransferase | C4-dicarboxylic acid, orotate and citrate transporter | galactitol-specific enzyme IIA component of PTS | galactitol-specific enzyme IIB component of PTS | galactitol-specific enzyme IIC component of PTS | D-tagatose 1,6-bisphosphate aldolase 2, subunit | lipoamide dehydrogenase, E3 component is part of three enzyme complexes | fused mannose-specific PTS enzymes: IIA component/IIB component | mannose-specific enzyme IIC component of PTS | mannose-specific enzyme IID component of PTS | methylglyoxal synthase | multifunctional nucleoside diphosphate kinase and apyrimidinic endonuclease and 3'-phosphodiesterase | NADH:ubiquinone oxidoreductase, membrane subunit A | NADH:ubiquinone oxidoreductase, chain B | NADH:ubiquinone oxidoreductase, chain C,D | NADH:ubiquinone oxidoreductase, chain E | NADH:ubiquinone oxidoreductase, chain F | NADH:ubiquinone oxidoreductase, chain G | NADH:ubiquinone oxidoreductase, membrane subunit H | NADH:ubiquinone oxidoreductase, chain I | NADH:ubiquinone oxidoreductase, membrane subunit J | NADH:ubiquinone oxidoreductase, membrane subunit K | NADH:ubiquinone oxidoreductase, membrane subunit L | NADH:ubiquinone oxidoreductase, membrane subunit M | nucleoside (except guanosine) transporter | polymyxin resistance protein B | phosphohistidinoprotein-hexose phosphotransferase component of PTS system (Hpr) | L-serine deaminase II | predicted serine transporter | succinate dehydrogenase, membrane subunit, binds cytochrome b556 | superoxide dismutase, Fe | pyridine nucleotide transhydrogenase, soluble | 2-oxoglutarate decarboxylase, thiamin-requiring | dihydrolipoyltranssuccinase | succinyl-CoA synthetase, NAD(P)-binding, alpha subunit | transketolase 1, thiamin-binding | nucleoside channel, receptor of phage T6 and colicin K | conserved protein | bifunctional aconitate hydratase 2/2-methylisocitrate dehydratase | ecotin, a serine protease inhibitor | fumarate reductase (anaerobic), membrane anchor subunit | galactitol-1-phosphate dehydrogenase, Zn-dependent and NAD(P)-binding | D-tagatose 1,6-bisphosphate aldolase 2, catalytic subunit | glucokinase | glycerol facilitator | glycerol kinase | heat shock protein | fused malic enzyme predicted oxidoreductase/predicted phosphotransacetylase | maltodextrin phosphorylase | 4-alpha-glucanotransferase (amylomaltase) | malate dehydrogenase, NAD(P)-binding | NADH:ubiquinone oxidoreductase, membrane subunit N | oligopeptide transporter subunit | phosphoenolpyruvate carboxykinase | aminopeptidase B | thiosulfate:cyanide sulfurtransferase (rhodanese) | PEP-protein phosphotransferase of PTS system (enzyme I) | fused D-ribose transporter subunits of ABC superfamily: ATP-binding components | D-ribose transporter subunit | predicted cytoplasmic sugar-binding protein | ribokinase | ribonucleoside hydrolase 1 | regulator of nucleoside diphosphate kinase | succinate dehydrogenase, flavoprotein subunit | succinate dehydrogenase, FeS subunit | 3-phosphoserine phosphatase | 3-mercaptopyruvate sulfurtransferase | succinyl-CoA synthetase, beta subunit | fused trehalose(maltose)-specific PTS enzyme: IIB component/IIC component | trehalose-6-P hydrolase | conserved inner membrane protein | predicted GTPase |
| 117 | 74 | 334 | 0.310 | 0.622 |  |  |  |  |  | b0038 b0037 b2091 b2096 b4517 b0154 b4037 b4180 b4054 b0223 b1036 b1110 b2700 b2681 b3103 b3293 b3662 b3698 b3954 b0464 b0064 b3210 b3237 b2143 b2782 b0155 b2592 b3816 b1200 b0014 b3540 b3673 b2369 b0757 b0756 b0758 b3868 b3869 b3925 b4143 b4142 b3867 b4350 b3526 b0891 b3938 b2924 b3600 b3601 b2926 b2144 b3917 b1916 b3590 b3244 b4324 b0156 b0631 b0892 b1969 b2145 b2471 b2450 b2560 b2958 b3082 b3083 b3358 b3602 b3713 b3937 b4394 b4395 b3427 | caiB caiC gatD gatY gnsA hemL malM rlmB tyrB yafJ ycdZ ycfJ ygaD ygaY yhaH yhdN nepI yidB yijO acrR araC arcB argR cdd chpA clcA clpB corA dhaK dnaK dppF emrD evgA galK galM galT glnG glnL glpX groL groS hemN hsdR kdgK lolA metJ mscS mtlD mtlR pgk sanA sbp sdiA selB tldD uxuR yadR ybeD rarA yedW yeiS yffB yffS yfhB yggN ygjM ygjN yhfK yibL yieF yiiX yjjX ytjC yzgL | crotonobetainyl CoA:carnitine CoA transferase | predicted crotonobetaine CoA ligase:carnitine CoA ligase | galactitol-1-phosphate dehydrogenase, Zn-dependent and NAD(P)-binding | D-tagatose 1,6-bisphosphate aldolase 2, catalytic subunit | predicted regulator of phosphatidylethanolamine synthesis | glutamate-1-semialdehyde aminotransferase (aminomutase) | maltose regulon periplasmic protein | 23S rRNA (Gm2251)-methyltransferase | tyrosine aminotransferase, tyrosine-repressible, PLP-dependent | predicted amidotransfease | predicted inner membrane protein | predicted protein | conserved protein | predicted transporter | predicted DNA-binding transcriptional regulator | DNA-binding transcriptional repressor | DNA-binding transcriptional dual regulator | hybrid sensory histidine kinase in two-component regulatory system with ArcA | DNA-binding transcriptional dual regulator, L-arginine-binding | cytidine/deoxycytidine deaminase | toxin of the ChpA-ChpR toxin-antitoxin system, endoribonuclease | chloride channel, voltage-gated | protein disaggregation chaperone | magnesium/nickel/cobalt transporter | dihydroxyacetone kinase, N-terminal domain | chaperone Hsp70, co-chaperone with DnaJ | dipeptide transporter | multidrug efflux system protein | DNA-binding response regulator in two-component regulatory system with EvgS | galactokinase | galactose-1-epimerase (mutarotase) | galactose-1-phosphate uridylyltransferase | fused DNA-binding response regulator in two-component regulatory system with GlnL: response regulator/sigma54 interaction protein | sensory histidine kinase in two-component regulatory system with GlnG | fructose 1,6-bisphosphatase II | Cpn60 chaperonin GroEL, large subunit of GroESL | Cpn10 chaperonin GroES, small subunit of GroESL | coproporphyrinogen III oxidase, SAM and NAD(P)H dependent, oxygen-independent | endonuclease R | ketodeoxygluconokinase | chaperone for lipoproteins | DNA-binding transcriptional repressor, S-adenosylmethionine-binding | mechanosensitive channel | mannitol-1-phosphate dehydrogenase, NAD(P)-binding | DNA-binding repressor | phosphoglycerate kinase | sulfate transporter subunit | DNA-binding transcriptional activator | selenocysteinyl-tRNA-specific translation factor | predicted peptidase | recombination protein | predicted DNA-binding response regulator in two-component system with YedV | CPZ-55 prophage; predicted protein | conserved inner membrane protein | chromate reductase, Class I, flavoprotein | predicted peptidoglycan peptidase | thiamin metabolism associated protein | phosphoglyceromutase 2, co-factor independent |
| 55 | 69 | 333 | 0.404 | 0.473 |  |  | 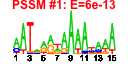 | 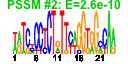 |  | b0512 b0618 b2368 b3264 b0535 b2883 b2542 b2539 b0139 b0343 b0076 b0383 b0530 b0531 b0534 b0533 b0193 b0164 b0385 b0539 b1115 b1471 b1576 b2109 b2123 b2230 b2272 b2354 b2408 b2444 b2445 b2577 b2578 b2629 b2633 b2778 b4282 b0258 b0300 b0301 b0304 b0305 b4534 b2550 b2624 b0140 b0342 b0254 b0136 b0137 b0138 b0218 b0358 b0363 b0375 b4579 b0603 b0690 b0964 b1503 b2338 b2503 b2534 b2630 b2631 b2761 b2882 b4464 b2940 | allB citC emrK envR fimZ guaD hcaD hcaF htrE lacY leuO phoA sfmA sfmC sfmF sfmH yaeF yaeI adrA ybcC ycfT yddK ydfD yehB yehR yfaA yfbM yfdK yfeN yffM yffN yfiE eamB yfjM yfjQ ygcG yjhE ykfC ykgA ykgB ykgC ykgD yphH alpA ecpD lacA perR yadK yadL yadM yafU yaiO yaiP yaiV yaiX ybdO yccT ydeR yfgF yfhR rnlA yfjO ygcB ygfO ygfQ yqgC | allantoinase | citrate lyase synthetase | EmrKY-TolC multidrug resistance efflux pump, membrane fusion protein component | DNA-binding transcriptional regulator | predicted DNA-binding transcriptional regulator | guanine deaminase | phenylpropionate dioxygenase, ferredoxin reductase subunit | 3-phenylpropionate dioxygenase, small (beta) subunit | predicted outer membrane usher protein | lactose/galactose transporter | DNA-binding transcriptional activator | bacterial alkaline phosphatase | predicted fimbrial-like adhesin protein | pilin chaperone, periplasmic | predicted lipoprotein | predicted phosphatase | predicted diguanylate cyclase | predicted inner membrane protein | Qin prophage; predicted protein | predicted outer membrane protein | conserved protein | predicted protein | CPS-53 (KpLE1) prophage; conserved protein | conserved outer membrane protein | CPZ-55 prophage; predicted protein | neutral amino-acid efflux system | CP4-57 prophage; predicted protein | conserved inner membrane protein | predicted oxidoreductase with FAD/NAD(P)-binding domain and dimerization domain | CP4-57 prophage; DNA-binding transcriptional activator | predicted periplasmic pilin chaperone | thiogalactoside acetyltransferase | CP4-6 prophage; predicted DNA-binding transcriptional regulator | predicted glucosyltransferase | predicted peptidase | CP4-57 prophage; RNase LS | conserved protein, member of DEAD box family | predicted transporter |
| 22 | 68 | 340 | 0.404 | 0.461 |  |  |  |  |  | b2064 b2132 b1711 b1736 b1738 b1874 b2065 b1961 b1594 b1612 b1611 b2436 b1192 b1613 b1159 b1424 b0826 b1399 b1400 b1278 b1713 b1609 b1294 b1290 b1272 b1429 b1430 b1274 b1610 b1618 b1960 b0766 b1219 b1220 b1268 b1321 b4529 b1383 b1419 b1434 b1435 b1438 b1647 b1643 b1770 b1773 b1787 b1799 b1908 b1929 b1930 b1976 b2173 b0835 b4532 b1820 b1858 b2010 b1329 b0813 b2176 b2011 b2170 b1641 b1644 b1645 b1786 b1798 | asmA bglX btuC chbA chbB cutC dcd dcm dgsA fumA fumC hemF ldcA manA mcrA mdoD moeB paaX paaY pgpB pheT rstB sapA sapF sohB tehA tehB topA tus uidR vsr ybhA ychN ychO yciQ ycjX ydbJ ydbL ydcA ydcN ydcP ydcQ ydhF ydhI ydjF ydjI yeaK yeaT yecA yedE yedF mtfA yeiR yliG yncN yobD znuC dacD mppA rhtA rtn sbcB setB slyB ydhJ ydhK yeaJ leuE | predicted assembly protein | beta-D-glucoside glucohydrolase, periplasmic | vitamin B12 transporter subunit: membrane component of ABC superfamily | N,N'-diacetylchitobiose-specific enzyme IIA component of PTS | N,N'-diacetylchitobiose-specific enzyme IIB component of PTS | copper homeostasis protein | 2'-deoxycytidine 5'-triphosphate deaminase | DNA cytosine methylase | DNA-binding transcriptional repressor | fumarate hydratase (fumarase A), aerobic Class I | fumarate hydratase (fumarase C),aerobic Class II | coproporphyrinogen III oxidase | L,D-carboxypeptidase A | mannose-6-phosphate isomerase | e14 prophage; 5-methylcytosine-specific restriction endonuclease B | glucan biosynthesis protein, periplasmic | molybdopterin synthase sulfurylase | DNA-binding transcriptional repressor of phenylacetic acid degradation, aryl-CoA responsive | predicted hexapeptide repeat acetyltransferase | phosphatidylglycerophosphatase B | phenylalanine tRNA synthetase, beta subunit | sensory histidine kinase in two-component regulatory system with RstA | predicted antimicrobial peptide transporter subunit | predicted inner membrane peptidase | potassium-tellurite ethidium and proflavin transporter | predicted S-adenosyl-L-methionine-dependent methyltransferase | DNA topoisomerase I, omega subunit | inhibitor of replication at Ter, DNA-binding protein | DNA mismatch endonuclease of very short patch repair | predicted hydrolase | conserved protein | predicted invasin | predicted inner membrane protein | conserved protein with nucleoside triphosphate hydrolase domain | predicted protein | predicted DNA-binding transcriptional regulator | predicted peptidase | predicted oxidoreductase | predicted aldolase | conserved metal-binding protein | predicted enzyme | predicted SAM-dependent methyltransferase | conserved inner membrane protein | zinc transporter subunit: ATP-binding component of ABC superfamily | D-alanyl-D-alanine carboxypeptidase (penicillin-binding protein 6b) | murein tripeptide (L-ala-gamma-D-glutamyl-meso-DAP) transporter subunit | threonine and homoserine efflux system | exonuclease I | lactose/glucose efflux system | outer membrane lipoprotein | undecaprenyl pyrophosphate phosphatase | predicted diguanylate cyclase | neutral amino-acid efflux system |
| 112 | 69 | 334 | 0.411 | 0.495 |  |  | 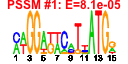 | 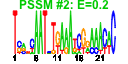 |  | b3974 b4316 b4207 b1905 b3560 b3559 b3867 b0851 b3724 b3726 b3725 b3727 b3728 b2620 b0850 b2471 b2618 b2619 b2296 b0463 b0462 b3131 b0159 b3570 b2901 b0437 b0623 b0733 b0734 b3860 b3177 b3176 b0200 b3385 b4480 b4349 b4348 b4000 b3617 b0439 b0222 b2813 b0211 b2684 b0578 b0134 b0133 b0143 b3616 b3384 b0490 b0735 b4515 b2605 b3248 b3407 b3697 b3714 b3764 b3865 b3858 b3866 b4022 b4251 b3146 b3147 b3148 b3149 b3150 | coaA fimC fklB ftnA glyQ glyS hemN nfsA phoU pstA pstB pstC pstS smpB ybjC yffB yfjF yfjG ackA acrA acrB agaR mtn bax bglA clpP cspE cydA cydB dsbA folP glmM gmhB gph hdfR hsdM hsdS hupA kbl lon lpcA mltA mltD mprA nfsB panB panC pcnB tdh trpS ybbL ybgE ybgT yfiB yhdE yhgF yidA yieG yifE yihA yihD yihI rluF yjgJ yraL yraM yraN diaA yraP | pantothenate kinase | chaperone, periplasmic | FKBP-type peptidyl-prolyl cis-trans isomerase (rotamase) | ferritin iron storage protein (cytoplasmic) | glycine tRNA synthetase, alpha subunit | glycine tRNA synthetase, beta subunit | coproporphyrinogen III oxidase, SAM and NAD(P)H dependent, oxygen-independent | nitroreductase A, NADPH-dependent, FMN-dependent | negative regulator of PhoR/PhoB two-component regulator | phosphate transporter subunit | trans-translation protein | predicted inner membrane protein | conserved protein | predicted protein | acetate kinase A and propionate kinase 2 | multidrug efflux system | multidrug efflux system protein | DNA-binding transcriptional dual regulator | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | 6-phospho-beta-glucosidase A | proteolytic subunit of ClpA-ClpP and ClpX-ClpP ATP-dependent serine proteases | DNA-binding transcriptional repressor | cytochrome d terminal oxidase, subunit I | cytochrome d terminal oxidase, subunit II | periplasmic protein disulfide isomerase I | 7,8-dihydropteroate synthase | phosphoglucosamine mutase | D,D-heptose 1,7-bisphosphate phosphatase | phosphoglycolate phosphatase | DNA-binding transcriptional regulator | DNA methylase M | specificity determinant for hsdM and hsdR | HU, DNA-binding transcriptional regulator, alpha subunit | glycine C-acetyltransferase | DNA-binding ATP-dependent protease La | D-sedoheptulose 7-phosphate isomerase | membrane-bound lytic murein transglycosylase A | predicted membrane-bound lytic murein transglycosylase D | DNA-binding transcriptional repressor of microcin B17 synthesis and multidrug efflux | dihydropteridine reductase, NAD(P)H-dependent, oxygen-insensitive | 3-methyl-2-oxobutanoate hydroxymethyltransferase | pantothenate synthetase | poly(A) polymerase I | threonine 3-dehydrogenase, NAD(P)-binding | tryptophanyl-tRNA synthetase | predicted transporter subunit: ATP-binding component of ABC superfamily | conserved inner membrane protein | predicted outer membrane lipoprotein | predicted transcriptional accessory protein | predicted hydrolase | GTP-binding protein | 23S rRNA pseudouridine synthase | predicted transcriptional regulator | predicted methyltransferase | DnaA initiator-associating factor for replication initiation |
| 93 | 49 | 337 | 0.462 | 0.535 |  |  |  |  |  | b2201 b2200 b2199 b2197 b2196 b2195 b2194 b1201 b0849 b1622 b2202 b1222 b1636 b1363 b1521 b1163 b1206 b1577 b1605 b1654 b1595 b4538 b0842 b0840 b1570 b1569 b1370 b1620 b1621 b1159 b0826 b1221 b1814 b1642 b1618 b1913 b4580 b0442 b0841 b1161 b1439 b1549 b1567 b1568 b1877 b2157 b2158 b1522 b4535 | ccmA ccmB ccmC ccmE ccmF ccmG ccmH dhaR grxA malY napC narX pdxY trkG uxaB ycgF ychM ydfE ydgI grxD ynfL yoeF cmr deoR dicA dicC insH malI malX mcrA moeB narL sdaA slyA uidR uvrC yaiT ybaV ybjG ycgX ydcR ydfO ydfW ydfX yecT yeiE yeiH yneF yniD | heme exporter subunit | periplasmic heme chaperone | heme lyase, CcmF subunit | periplasmic thioredoxin of cytochrome c-type biogenesis | heme lyase, CcmH subunit | predicted DNA-binding transcriptional regulator, dihydroxyacetone | glutaredoxin 1, redox coenzyme for ribonucleotide reductase (RNR1a) | bifunctional beta-cystathionase, PLP-dependent/ regulator of maltose regulon | nitrate reductase, cytochrome c-type, periplasmic | sensory histidine kinase in two-component regulatory system with NarL | pyridoxal kinase 2/pyridoxine kinase | Rac prophage; potassium transporter subunit | altronate oxidoreductase, NAD-dependent | predicted FAD-binding phosphodiesterase | predicted transporter | predicted arginine/ornithine antiporter transporter | conserved protein | predicted DNA-binding transcriptional regulator | multidrug efflux system protein | DNA-binding transcriptional repressor | Qin prophage; predicted regulator for DicB | Qin prophage; DNA-binding transcriptional regulator for DicB | IS5 transposase and trans-activator | fused maltose and glucose-specific PTS enzymes: IIB component -! IIC component | e14 prophage; 5-methylcytosine-specific restriction endonuclease B | molybdopterin synthase sulfurylase | DNA-binding response regulator in two-component regulatory system with NarX (or NarQ) | L-serine deaminase I | DNA-binding transcriptional activator | excinuclease UvrABC, endonuclease subunit | undecaprenyl pyrophosphate phosphatase | predicted protein | fused predicted DNA-binding transcriptional regulator/predicted amino transferase | Qin prophage; predicted protein | conserved inner membrane protein | predicted diguanylate cyclase |
| 37 | 61 | 342 | 0.508 | 0.499 |  |  |  |  |  | b3139 b3141 b0505 b3501 b2460 b0509 b2831 b4106 b4101 b4100 b4099 b4098 b4096 b0241 b0400 b2987 b0397 b2879 b2881 b0013 b0011 b0137 b0138 b0326 b0504 b0544 b0787 b1001 b2427 b2654 b2919 b2878 b2880 b2882 b2983 b2986 b3027 b4027 b4249 b4358 b0235 b1149 b2382 b2650 b4211 b0035 b0622 b2709 b2710 b0399 b0236 b3903 b0398 b1157 b0141 b4526 b1153 b2649 b2658 b2915 b4212 | agaC agaI allA arsR eutQ glxR mutH phnC phnG phnH phnI phnJ phnL phoE phoR pitB sbcC ssnA xdhD yaaI yaaW yadL yadM yahL allS ybcK ybhM yccE yfeT scpB ygfK ygfM ygfO yghQ yghT ygiZ yjbF yjgI yjjN ykfJ ymfN ypdC ypjC ytfG caiE pagP norR norV phoB prfH rhaA sbcD stfE yadN ydaE ymfQ yqfE ytfH | N-acetylgalactosamine-specific enzyme IIC component of PTS | galactosamine-6-phosphate isomerase | ureidoglycolate hydrolase | DNA-binding transcriptional repressor | conserved protein | tartronate semialdehyde reductase, NADH-dependent | methyl-directed mismatch repair protein | phosphonate/organophosphate ester transporter subunit | carbon-phosphorus lyase complex subunit | outer membrane phosphoporin protein E | sensory histidine kinase in two-component regulatory system with PhoB | phosphate transporter | exonuclease, dsDNA, ATP-dependent | predicted chlorohydrolase/aminohydrolase | fused predicted xanthine/hypoxanthine oxidase: molybdopterin-binding subunit/Fe-S binding subunit | predicted protein | predicted fimbrial-like adhesin protein | DNA-binding transcriptional activator of the allD operon | DLP12 prophage; predicted recombinase | conserved inner membrane protein | predicted DNA-binding transcriptional regulator | methylmalonyl-CoA decarboxylase, biotin-independent | predicted oxidoreductase, Fe-S subunit | predicted oxidoreductase | predicted transporter | predicted inner membrane protein | predicted protein with nucleoside triphosphate hydrolase domain | predicted lipoprotein | predicted oxidoreductase with NAD(P)-binding Rossmann-fold domain | predicted oxidoreductase, Zn-dependent and NAD(P)-binding | e14 prophage; predicted DNA-binding transcriptional regulator | predicted DNA-binding protein | NAD(P)H:quinone oxidoreductase | predicted acyl transferase | palmitoyl transferase for Lipid A | DNA-binding transcriptional activator | flavorubredoxin oxidoreductase | DNA-binding response regulator in two-component regulatory system with PhoR (or CreC) | L-rhamnose isomerase | Rac prophage; conserved protein | e14 prophage; conserved protein | predicted transcriptional regulator |
| 96 | 77 | 340 | 0.519 | 0.538 |  |  |  | 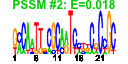 |  | b0463 b0462 b0464 b3131 b3210 b4398 b4137 b2927 b3406 b0104 b2813 b2684 b0196 b0025 b3247 b0098 b3407 b3554 b3602 b3697 b3858 b3921 b4022 b4146 b3146 b3147 b3148 b3150 b2786 b3533 b3536 b3537 b3538 b3531 b0103 b4397 b3182 b0092 b3075 b0080 b0094 b0089 b0148 b0082 b0087 b0081 b3226 b3405 b2821 b2822 b2784 b4396 b3987 b3988 b2785 b0056 b0122 b0102 b0101 b0391 b0482 b0483 b2683 b2682 b2929 b3233 b3252 b3534 b3535 b3920 b4295 b4403 b2900 b3095 b4220 b4221 b2412 | acrA acrB acrR agaR arcB creB cutA epd greB guaC mltA mprA rcsF ribF rng secA yhgF yiaF yibL yidA yihD yiiR rluF yjeK yraL yraM yraN yraP barA bcsA bcsE bcsF bcsG bcsZ coaE creA dacB ddlB ebgR fruR ftsA ftsW hrpB mraW mraY mraZ nanR ompR ptrA recC relA rob rpoB rpoC rumA yacC yacF yacG yaiE ybaP ybaQ ygaH ygaZ yggD yhcB csrD yhjQ yhjR yiiQ yjhU yjtD yqfB yqjA ytfM ytfN zipA | multidrug efflux system | multidrug efflux system protein | DNA-binding transcriptional repressor | DNA-binding transcriptional dual regulator | hybrid sensory histidine kinase in two-component regulatory system with ArcA | DNA-binding response regulator in two-component regulatory system with CreC | copper binding protein, copper sensitivity | D-erythrose 4-phosphate dehydrogenase | transcription elongation factor | GMP reductase | membrane-bound lytic murein transglycosylase A | DNA-binding transcriptional repressor of microcin B17 synthesis and multidrug efflux | predicted outer membrane protein, signal | bifunctional riboflavin kinase/FAD synthetase | ribonuclease G | preprotein translocase subunit, ATPase | predicted transcriptional accessory protein | conserved protein | predicted hydrolase | conserved inner membrane protein | 23S rRNA pseudouridine synthase | predicted lysine aminomutase | predicted methyltransferase | predicted protein | hybrid sensory histidine kinase, in two-component regulatory system with UvrY | cellulose synthase, catalytic subunit | predicted inner membrane protein | endo-1,4-D-glucanase | dephospho-CoA kinase | D-alanyl-D-alanine carboxypeptidase | D-alanine:D-alanine ligase | ATP-binding cell division protein involved in recruitment of FtsK to Z ring | integral membrane protein involved in stabilizing FstZ ring during cell division | predicted ATP-dependent helicase | S-adenosyl-dependent methyltransferase activity on membrane-located substrates | phospho-N-acetylmuramoyl-pentapeptide transferase | DNA-binding response regulator in two-component regulatory system with EnvZ | protease III | exonuclease V (RecBCD complex), gamma chain | (p)ppGpp synthetase I/GTP pyrophosphokinase | DNA-binding transcriptional activator | RNA polymerase, beta subunit | RNA polymerase, beta prime subunit | 23S rRNA (uracil-5)-methyltransferase | predicted DNA-binding transcriptional regulator | predicted transporter | KpLE2 phage-like element; predicted DNA-binding transcriptional regulator | predicted rRNA methyltransferase | predicted outer membrane protein and surface antigen | cell division protein involved in Z ring assembly |
| 75 | 60 | 345 | 0.550 | 0.545 |  |  | 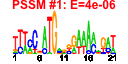 | 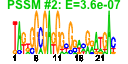 |  | b0050 b0159 b3162 b0055 b4361 b0173 b0048 b0172 b3178 b3425 b3424 b3423 b3181 b0125 b0054 b0051 b0459 b0635 b0052 b3018 b3164 b3622 b3988 b3179 b1916 b0053 b2516 b2517 b2512 b2583 b2765 b3606 b3866 b4360 b2689 b2690 b3966 b3388 b0015 b2525 b0890 b0059 b4175 b2526 b3618 b3028 b3249 b3982 b3961 b4155 b3363 b3067 b4425 b0269 b0822 b2524 b4461 b3180 b3467 b3468 | apaG mtn deaD djlA dnaC dxr folA frr hflB glpE glpG glpR greA hpt imp ksgA maa mrdA pdxA plsC pnp rfaL rpoC rrmJ sdiA surA yfgA yfgB yfgL yfiP sscR yibK yihI yjjA yqaA yqaB btuB damX dnaJ fdx ftsK hepA hflC hscA htrL mdaB mreD nusG oxyR poxA ppiA rpoD yagF ybiV iscX yfjD yhbY yhhM yhhN | protein associated with Co2+ and Mg2+ efflux | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | ATP-dependent RNA helicase | DnaJ-like protein, membrane anchored | DNA biosynthesis protein | 1-deoxy-D-xylulose 5-phosphate reductoisomerase | dihydrofolate reductase | ribosome recycling factor | protease, ATP-dependent zinc-metallo | thiosulfate:cyanide sulfurtransferase (rhodanese) | predicted intramembrane serine protease | DNA-binding transcriptional repressor | transcription elongation factor | hypoxanthine phosphoribosyltransferase | exported protein required for envelope biosynthesis and integrity | S-adenosylmethionine-6-N',N'-adenosyl (rRNA) dimethyltransferase | maltose O-acetyltransferase | transpeptidase involved in peptidoglycan synthesis (penicillin-binding protein 2) | 4-hydroxy-L-threonine phosphate dehydrogenase, NAD-dependent | 1-acyl-sn-glycerol-3-phosphate acyltransferase | polynucleotide phosphorylase/polyadenylase | O-antigen ligase | RNA polymerase, beta prime subunit | 23S rRNA methyltransferase | DNA-binding transcriptional activator | peptidyl-prolyl cis-trans isomerase (PPIase) | conserved protein | predicted enzyme | protein assembly complex, lipoprotein component | 6-pyruvoyl tetrahydrobiopterin synthase (PTPS) | predicted rRNA methylase | conserved inner membrane protein | predicted hydrolase | vitamin B12/cobalamin outer membrane transporter | predicted protein | chaperone Hsp40, co-chaperone with DnaK | [2Fe-2S] ferredoxin | DNA-binding membrane protein required for chromosome resolution and partitioning | RNA polymerase-associated helicase protein (ATPase and RNA polymerase recycling factor) | modulator for HflB protease specific for phage lambda cII repressor | DnaK-like molecular chaperone specific for IscU | NADPH quinone reductase | cell wall structural complex MreBCD transmembrane component MreD | transcription termination factor | DNA-binding transcriptional dual regulator | predicted lysyl-tRNA synthetase | peptidyl-prolyl cis-trans isomerase A (rotamase A) | RNA polymerase, sigma 70 (sigma D) factor | CP4-6 prophage; predicted dehydratase | predicted inner membrane protein | predicted RNA-binding protein |
| 16 | 83 | 334 | 0.573 | 0.550 |  |  |  |  |  | b0114 b0115 b1241 b2782 b0438 b0638 b3357 b3388 b1198 b1200 b1199 b0015 b0014 b2779 b2925 b0904 b0239 b0084 b0890 b0083 b0095 b0757 b0756 b2688 b0369 b2753 b2622 b0891 b2781 b3028 b0091 b0088 b0085 b0086 b0090 b0677 b0676 b0675 b2082 b0425 b0237 b3916 b0902 b0903 b4025 b2297 b2780 b2820 b0797 b0641 b3067 b0008 b3919 b3384 b0101 b0225 b0482 b0483 b0492 b0493 b4515 b0794 b0795 b0792 b0793 b0822 b2017 b2615 b2628 b2630 b2631 b2862 b3157 b3356 b4539 b1852 b0381 b2837 b0238 b3055 b1380 b0796 b2380 | aceE aceF adhE chpA clpX cobC crp damX dhaM dhaK dhaL dnaJ dnaK eno fbaA focA frsA ftsI ftsK ftsL ftsZ galK galM gshA hemB iap intA lolA mazG mdaB murC murD murE murF murG nagA nagC nagD ogrK panE pepD pfkA pflA pflB pgi pta pyrG recB rhlE rlpB rpoD talB tpiA trpS yacG yafQ ybaP ybaQ ybbN ybbO ybgT ybhF ybhG ybhR ybhS ybiV yefM nadK yfjL rnlA yfjO yhbT yhfA yoeB zwf ddlA galR gpt htrG ldhA ybiH ypdA | pyruvate dehydrogenase, decarboxylase component E1, thiamin-binding | pyruvate dehydrogenase, dihydrolipoyltransacetylase component E2 | fused acetaldehyde-CoA dehydrogenase/iron-dependent alcohol dehydrogenase/pyruvate-formate lyase deactivase | toxin of the ChpA-ChpR toxin-antitoxin system, endoribonuclease | ATPase and specificity subunit of ClpX-ClpP ATP-dependent serine protease | predicted alpha-ribazole-5'-P phosphatase | DNA-binding transcriptional dual regulator | predicted protein | fused predicted dihydroxyacetone-specific PTS enzymes: HPr component/EI component | dihydroxyacetone kinase, N-terminal domain | dihydroxyacetone kinase, C-terminal domain | chaperone Hsp40, co-chaperone with DnaK | chaperone Hsp70, co-chaperone with DnaJ | enolase | fructose-bisphosphate aldolase, class II | formate transporter | hydrolase, binds to enzyme IIA(Glc) | transpeptidase involved in septal peptidoglycan synthesis (penicillin-binding protein 3) | DNA-binding membrane protein required for chromosome resolution and partitioning | membrane bound cell division protein at septum containing leucine zipper motif | GTP-binding tubulin-like cell division protein | galactokinase | galactose-1-epimerase (mutarotase) | gamma-glutamate-cysteine ligase | porphobilinogen synthase | aminopeptidase in alkaline phosphatase isozyme conversion | CP4-57 prophage; integrase | chaperone for lipoproteins | nucleoside triphosphate pyrophosphohydrolase | NADPH quinone reductase | UDP-N-acetylmuramate:L-alanine ligase | UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate:meso- diaminopimelate ligase | UDP-N-acetylmuramoyl-tripeptide:D-alanyl-D- alanine ligase | N-acetylglucosaminyl transferase | N-acetylglucosamine-6-phosphate deacetylase | DNA-binding transcriptional dual regulator, repressor of N-acetylglucosamine | UMP phosphatase | DNA-binding transcriptional regulator prophage P2 remnant | 2-dehydropantoate reductase, NADPH-specific | aminoacyl-histidine dipeptidase (peptidase D) | 6-phosphofructokinase I | pyruvate formate lyase activating enzyme 1 | pyruvate formate lyase I | glucosephosphate isomerase | phosphate acetyltransferase | CTP synthetase | exonuclease V (RecBCD complex), beta subunit | RNA helicase | minor lipoprotein | RNA polymerase, sigma 70 (sigma D) factor | transaldolase B | triosephosphate isomerase | tryptophanyl-tRNA synthetase | conserved protein | predicted toxin of the YafQ-DinJ toxin-antitoxin system | predicted DNA-binding transcriptional regulator | predicted thioredoxin domain-containing protein | predicted oxidoreductase with NAD(P)-binding Rossmann-fold domain | fused predicted transporter subunits of ABC superfamily: ATP-binding components | predicted membrane fusion protein (MFP) component of efflux pump, membrane anchor | predicted transporter subunit: membrane component of ABC superfamily | predicted hydrolase | antitoxin of the YoeB-YefM toxin-antitoxin system | NAD kinase | CP4-57 prophage; predicted protein | CP4-57 prophage; RNase LS | predicted lipid carrier protein | toxin of the YoeB-YefM toxin-antitoxin system | glucose-6-phosphate dehydrogenase | D-alanine-D-alanine ligase A | DNA-binding transcriptional repressor | guanine-hypoxanthine phosphoribosyltransferase | predicted signal transduction protein (SH3 domain) | fermentative D-lactate dehydrogenase, NAD-dependent | predicted sensory kinase in two-component system with YpdB |
| 83 | 78 | 343 | 0.613 | 0.486 |  |  |  |  |  | b3412 b3918 b3807 b3066 b2945 b2798 b3891 b4291 b4290 b4287 b0356 b0355 b0357 b3500 b3401 b3400 b3931 b4359 b4118 b3250 b3635 b3163 b3183 b3019 b3498 b0405 b3822 b3778 b3461 b2944 b3747 b3706 b2832 b2955 b4469 b3084 b3184 b3352 b3459 b3497 b3499 b3527 b3529 b3539 b3552 b4049 b4176 b3399 b3475 b0313 b3634 b3162 b4289 b4288 b3178 b3933 b0759 b3741 b3740 b0475 b3687 b3686 b4119 b4242 b3821 b3775 b3794 b3247 b3643 b3179 b4371 b3364 b2765 b4470 b3606 b4001 b2845 b3398 | bioH cdh cyaY dnaG endA xni fdhE fecA fecB fecE frmA frmB frmR gor hslO hslR hslU mdoB melR mreC mutM nlpI obgE parC prlC queA recQ rep rpoH yggI kup mnmE ygdQ yggW ygiQ rlmG yhbE yheS yhhK yhiQ yhiR yhjJ yhjK yhjV yiaD dusA yjeT yrfG acpT betI coaD deaD fecC fecD hflB ftsN galE mnmG gidB hemH ibpA ibpB melA mgtA pldA ppiC rffM rng rph rrmJ rsmC tsgA sscR yhaM yibK yjaH yqeG yrfF | carboxylesterase of pimeloyl-CoA synthesis | CDP-diacylglycerol phosphotidylhydrolase | frataxin, iron-binding and oxidizing protein | DNA primase | DNA-specific endonuclease I | exonuclease IX (5'-3' exonuclease) | formate dehydrogenase formation protein | KpLE2 phage-like element; ferric citrate outer membrane transporter | KpLE2 phage-like element; iron-dicitrate transporter subunit | alcohol dehydrogenase class III/glutathione-dependent formaldehyde dehydrogenase | predicted esterase | regulator protein that represses frmRAB operon | glutathione oxidoreductase | heat shock protein Hsp33 | ribosome-associated heat shock protein Hsp15 | molecular chaperone and ATPase component of HslUV protease | phosphoglycerol transferase I | DNA-binding transcriptional dual regulator | cell wall structural complex MreBCD transmembrane component MreC | formamidopyrimidine/5-formyluracil/ 5-hydroxymethyluracil DNA glycosylase | conserved protein | GTPase involved in cell partioning and DNA repair | DNA topoisomerase IV, subunit A | oligopeptidase A | S-adenosylmethionine:tRNA ribosyltransferase-isomerase | ATP-dependent DNA helicase | DNA helicase and single-stranded DNA-dependent ATPase | RNA polymerase, sigma 32 (sigma H) factor | potassium transporter | GTPase | predicted inner membrane protein | predicted oxidoreductase | predicted methyltransferase small domain | conserved inner membrane protein | fused predicted transporter subunits of ABC superfamily: ATP-binding components | predicted SAM-dependent methyltransferase | predicted DNA (exogenous) processing protein | predicted zinc-dependent peptidase | predicted diguanylate cyclase | predicted transporter | predicted outer membrane lipoprotein | tRNA-dihydrouridine synthase A | predicted hydrolase | holo-(acyl carrier protein) synthase 2 | DNA-binding transcriptional repressor | pantetheine-phosphate adenylyltransferase | ATP-dependent RNA helicase | protease, ATP-dependent zinc-metallo | essential cell division protein | UDP-galactose-4-epimerase | glucose-inhibited cell-division protein | methyltransferase, SAM-dependent methyltransferase, glucose-inhibited cell-division protein | ferrochelatase | heat shock chaperone | alpha-galactosidase, NAD(P)-binding | magnesium transporter | outer membrane phospholipase A | peptidyl-prolyl cis-trans isomerase C (rotamase C) | UDP-N-acetyl-D-mannosaminuronic acid transferase | ribonuclease G | defective ribonuclease PH | 23S rRNA methyltransferase | 16S rRNA m2G1207 methylase | 6-pyruvoyl tetrahydrobiopterin synthase (PTPS) | predicted rRNA methylase |
| 41 | 67 | 339 | 0.613 | 0.515 |  |  |  |  |  | b0376 b2714 b1992 b1991 b1993 b0624 b1954 b0879 b0394 b0730 b0760 b0243 b1305 b1564 b1563 b0377 b0208 b0209 b0378 b0466 b0580 b0815 b0847 b0858 b0898 b0956 b0947 b0959 b1128 b1162 b1446 b1477 b1962 b2006 b2304 b2623 b0457 b0515 b0833 b0834 b0952 b2136 b0752 b0111 b1507 b0537 b0465 b0403 b0761 b1715 b1304 b1306 b1307 b1303 b0210 b0444 b0762 b0844 b0845 b0909 b1799 b2063 b2102 b2244 b1145 b4532 b1793 | ampH ascG cobS cobT cobU crcB macB mak mngR modF proA pspB relB relE sbmA yafC yafD yaiW ybaM ybdJ ybiP ybjL ybjO ycaD ycbG ycbX sxy ycfD ycgE ydcY yddM yedJ yeeW yfcH yfjH ylaB ylbA yliE yliF ymbA yohD zitB ampE hipA intD kefA malZ modE pheM pspA pspC pspD pspF yafE queC ybhT ybjI ybjJ ycaL yeaT yegH yegX yfaD ymfK yncN yoaF | beta-lactamase/D-alanine carboxypeptidase | DNA-binding transcriptional repressor | cobalamin 5'-phosphate synthase | nicotinate-nucleotide dimethylbenzimidazole-P phophoribosyl transferase | bifunctional cobinamide kinase/ cobinamide phosphate guanylyltransferase | predicted inner membrane protein associated with chromosome condensation | fused macrolide transporter subunits of ABC superfamily: ATP-binding component/membrane component | manno(fructo)kinase | DNA-binding transcriptional dual regulator, fatty-acyl-binding | fused molybdate transporter subunits of ABC superfamily: ATP-binding components | gamma-glutamylphosphate reductase | DNA-binding transcriptional regulator of psp operon | Qin prophage; bifunctional antitoxin of the RelE-RelB toxin-antitoxin system/ transcriptional repressor | Qin prophage; toxin of the RelE-RelB toxin-antitoxin system | predicted transporter | predicted DNA-binding transcriptional regulator | conserved protein | predicted protein | predicted inner membrane protein | predicted hydrolase, inner membrane | predicted 2Fe-2S cluster-containing protein | predicted phosphohydrolase | conserved protein with NAD(P)-binding Rossmann-fold domain | CP4-57 prophage; predicted protein | conserved inner membrane protein | predicted diguanylate cyclase | zinc efflux system | regulator with hipB | DLP12 prophage; predicted integrase | fused conserved protein | maltodextrin glucosidase | DNA-binding transcriptional dual regulator | phenylalanyl-tRNA synthetase operon leader peptide | regulatory protein for phage-shock-protein operon | DNA-binding transcriptional activator | peripheral inner membrane phage-shock protein | predicted S-adenosyl-L-methionine-dependent methyltransferase | predicted aluminum resistance protein | predicted peptidase with chaperone function | fused predicted membrane protein/predicted membrane protein | predicted hydrolase | e14 prophage; repressor protein phage e14 | conserved outer membrane protein |
| 119 | 78 | 341 | 0.615 | 0.497 |  |  |  |  |  | b4117 b4085 b4366 b4131 b4132 b3690 b3717 b4478 b0799 b3895 b4317 b4318 b4319 b3898 b4498 b3681 b3558 b4283 b4345 b4120 b3856 b3935 b3593 b3482 b3188 b3583 b4449 b0958 b4194 b4198 b4192 b0322 b0798 b0800 b2858 b2921 b3043 b3085 b3086 b3411 b3483 b3488 b3490 b3520 b3561 b3584 b3585 b3594 b3596 b3595 b4555 b3689 b3716 b3765 b4482 b3862 b3889 b3890 b4050 b4130 b4145 b4157 b4158 b4274 b4281 b4282 b4308 b4333 b4486 b2874 b2941 b3047 b3050 b3051 b3443 b4552 b4553 b3808 | adiA alsE bglJ cadA cadB cbrA cbrC dgoD dinG fdhD fimD fimF fimG frvX gatR insK mcrC melB mobB priA rhsA rhsB sfsB sgbE sulA ulaB ulaF ulaG ybiA ybiB ygeN ygfI ygiL ygjP ygjQ yhgA yhhH yhiJ yhjB wecH yiaT yiaU yibA yibG yibJ yicS yidR cbrB yifB yigE yihG yiiE yiiF pspG yjdL yjeJ yjeN yjeO yjhD yjhE yjhR yjiK yjiV yqeA yqgD yqiH yqiJ yqiK yrhA yrhC ysaB | biodegradative arginine decarboxylase | allulose-6-phosphate 3-epimerase | DNA-binding transcriptional activator | lysine decarboxylase 1 | predicted lysine/cadaverine transporter | predicted oxidoreductase with FAD/NAD(P)-binding domain | conserved protein | galactonate dehydratase | ATP-dependent DNA helicase | formate dehydrogenase formation protein | outer membrane usher protein, type 1 fimbrial synthesis | minor component of type 1 fimbriae | predicted endo-1,4-beta-glucanase | IS150 conserved protein InsB | 5-methylcytosine-specific restriction enzyme McrBC, subunit McrC | melibiose:sodium symporter | molybdopterin-guanine dinucleotide biosynthesis protein B | Primosome factor n' (replication factor Y) | rhsA element core protein RshA | rhsB element core protein RshB | DNA-binding transcriptional activator of maltose metabolism | L-ribulose-5-phosphate 4-epimerase | SOS cell division inhibitor | L-ascorbate-specific enzyme IIB component of PTS | L-ribulose 5-phosphate 4-epimerase | predicted L-ascorbate 6-phosphate lactonase | predicted transferase/phosphorylase | predicted DNA-binding transcriptional regulator | predicted fimbrial-like adhesin protein | predicted metal dependent hydrolase | predicted thioredoxin-like protein | predicted transposase | predicted protein | predicted DNA-binding response regulator in two-component regulatory system | conserved inner membrane protein | lyase containing HEAT-repeat | predicted inner membrane protein | predicted bifunctional enzyme and transcriptional regulator | predicted endonuclease | predicted transcriptional regulator | phage shock protein G | predicted transporter | predicted amino acid kinase | predicted periplasmic pilin chaperone |
| 138 | 80 | 335 | 0.674 | 0.521 |  |  | 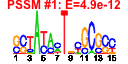 | 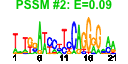 |  | b2317 b1850 b1851 b2685 b1095 b0634 b2234 b2235 b0472 b3052 b0633 b0471 b0636 b2236 b2299 b2300 b3871 b3974 b4361 b0470 b2344 b3058 b4517 b2496 b0369 b0154 b0473 b1531 b1530 b3930 b2264 b3396 b0635 b2733 b0851 b3258 b3724 b2259 b3259 b3726 b3725 b3727 b3728 b2297 b4177 b1676 b0797 b2567 b1084 b0911 b2797 b2796 b2937 b2576 b2533 b0503 b0685 b0850 b1036 b1660 b1825 b1915 b2174 b2263 b2398 b2517 b2562 b2583 b2618 b2619 b2700 b3064 b3257 b4360 b4391 b0458 b1826 b2791 b2792 b2949 | dedA eda edd emrA fabF mrdB nrdA nrdB recR rfaE rlpA ybaB ybeA yfaE yfcD yfcE typA coaA dnaC dnaX fadL folB gnsA hda hemB hemL htpG marA marR menA menD mrcA mrdA mutS nfsA panF phoU pmrD prmA pstA pstB pstC pstS pta purA pykF rhlE rnc rne rpsA sdaB sdaC speB srmB suhB ybbB ybfE ybjC ycdZ ydhC yebO yecF yeiU yfbB yfeC yfgB yfhL yfiP yfjF yfjG ygaD ygjD yhdT yjjA yjjK ylaC mgrB truC yqcC yqgF | conserved inner membrane protein | multifunctional 2-keto-3-deoxygluconate 6-phosphate aldolase and 2-keto-4-hydroxyglutarate aldolase and oxaloacetate decarboxylase | 6-phosphogluconate dehydratase | multidrug efflux system | 3-oxoacyl-[acyl-carrier-protein] synthase II | cell wall shape-determining protein | ribonucleoside diphosphate reductase 1, alpha subunit | ribonucleoside diphosphate reductase 1, beta subunit, ferritin-like protein | gap repair protein | fused heptose 7-phosphate kinase/heptose 1-phosphate adenyltransferase | minor lipoprotein | conserved protein | predicted 2Fe-2S cluster-containing protein | predicted NUDIX hydrolase | predicted phosphatase | GTP-binding protein | pantothenate kinase | DNA biosynthesis protein | DNA polymerase III/DNA elongation factor III, tau and gamma subunits | long-chain fatty acid outer membrane transporter | bifunctional dihydroneopterin aldolase/dihydroneopterin triphosphate 2'-epimerase | predicted regulator of phosphatidylethanolamine synthesis | ATPase regulatory factor involved in DnaA inactivation | porphobilinogen synthase | glutamate-1-semialdehyde aminotransferase (aminomutase) | molecular chaperone HSP90 family | DNA-binding transcriptional dual activator of multiple antibiotic resistance | DNA-binding transcriptional repressor of multiple antibiotic resistance | 1,4-dihydroxy-2-naphthoate octaprenyltransferase | bifunctional 2-oxoglutarate decarboxylase/ SHCHC synthase | fused penicillin-binding protein 1a: murein transglycosylase/murein transpeptidase | transpeptidase involved in peptidoglycan synthesis (penicillin-binding protein 2) | methyl-directed mismatch repair protein | nitroreductase A, NADPH-dependent, FMN-dependent | pantothenate:sodium symporter | negative regulator of PhoR/PhoB two-component regulator | polymyxin resistance protein B | methylase for 50S ribosomal subunit protein L11 | phosphate transporter subunit | phosphate acetyltransferase | adenylosuccinate synthetase | pyruvate kinase I | RNA helicase | RNase III | fused ribonucleaseE: endoribonuclease/RNA-binding protein/RNA degradosome binding protein | 30S ribosomal subunit protein S1 | L-serine deaminase II | predicted serine transporter | agmatinase | ATP-dependent RNA helicase | inositol monophosphatase | tRNA 2-selenouridine synthase, selenophosphate-dependent | lexA-regulated predicted protein | predicted inner membrane protein | predicted transporter | predicted protein | undecaprenyl pyrophosphate phosphatase | predicted peptidase | predicted DNA-binding transcriptional regulator | predicted enzyme | predicted 4Fe-4S cluster-containing protein | fused predicted transporter subunits of ABC superfamily: ATP-binding components | tRNA pseudouridine synthase | predicted Holliday junction resolvase |
| 65 | 79 | 334 | 0.698 | 0.550 |  |  | 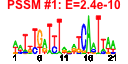 | 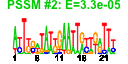 |  | b3242 b0034 b4123 b4154 b4153 b4151 b4122 b2977 b2997 b2713 b3942 b3222 b3367 b4021 b3756 b0201 b0280 b2343 b2447 b2448 b2450 b3156 b3221 b3820 b4128 b4559 b4252 b4279 b4280 b0564 b0620 b3368 b0904 b3128 b3124 b3126 b3127 b3125 b3214 b2988 b0281 b4435 b3909 b2289 b4019 b3477 b3480 b3481 b0903 b3854 b0556 b2405 b0382 b0559 b1675 b1963 b2001 b2443 b2446 b2449 b2579 b2670 b2754 b2755 b2756 b2757 b2758 b2759 b2760 b3157 b3159 b3253 b3573 b4253 b4256 b4326 b4380 b4379 b4533 | aaeX caiF dcuB frdA frdB frdD fumB glcG hybO hydN katG nanK nirC pepE rrsC rrsH yagN yfcZ yffP yffQ yffS yhbS yhcH yigI yjdK yjdO yjgK yjhB yjhC appY dpiA cysG focA garD garK garL garP garR gltF gsp intF kdgT lrhA metH nikB nikE nikR pflB rrlA rzpD xapR iraP ybcW ydhZ yedR yeeR yffL yffO yffR yfiD ygaW ygbF ygbT ygcH ygcI ygcJ ygcK ygcL yhbT yhbV yhdH ysaA yjgL yjgM yjiD yjjI yjjW ynfO | membrane protein of efflux system | DNA-binding transcriptional activator | C4-dicarboxylate antiporter | fumarate reductase (anaerobic) catalytic and NAD/flavoprotein subunit | fumarate reductase (anaerobic), Fe-S subunit | fumarate reductase (anaerobic), membrane anchor subunit | anaerobic class I fumarate hydratase (fumarase B) | conserved protein | hydrogenase 2, small subunit | formate dehydrogenase-H, [4Fe-4S] ferredoxin subunit | catalase/hydroperoxidase HPI(I) | predicted N-acetylmannosamine kinase | nitrite transporter | (alpha)-aspartyl dipeptidase | 16S ribosomal RNA of rrnC operon | 16S ribosomal RNA of rrnH operon | CP4-6 prophage; predicted protein | CPZ-55 prophage; predicted protein | predicted acyltransferase with acyl-CoA N-acyltransferase domain | predicted protein | KpLE2 phage-like element; predicted transporter | KpLE2 phage-like element; predicted oxidoreductase | DLP12 prophage; DNA-binding transcriptional activator | DNA-binding response regulator in two-component regulatory system with citA | fused siroheme synthase 1,3-dimethyluroporphyriongen III dehydrogenase and siroheme ferrochelatase/uroporphyrinogen methyltransferase | formate transporter | (D)-galactarate dehydrogenase | glycerate kinase I | alpha-dehydro-beta-deoxy-D-glucarate aldolase | predicted (D)-galactarate transporter | tartronate semialdehyde reductase | periplasmic protein | fused glutathionylspermidine amidase/glutathionylspermidine synthetase | CP4-6 prophage; predicted phage integrase | 2-keto-3-deoxy-D-gluconate transporter | DNA-binding transcriptional repressor of flagellar, motility and chemotaxis genes | homocysteine-N5-methyltetrahydrofolate transmethylase, B12-dependent | nickel transporter subunit | DNA-binding transcriptional repressor, Ni-binding | pyruvate formate lyase I | 23S ribosomal RNA of rrnA operon | DLP12 prophage; predicted murein endopeptidase | DLP12 prophage; predicted protein | predicted inner membrane protein | CP4-44 prophage; predicted membrane protein | pyruvate formate lyase subunit | predicted lipid carrier protein | predicted protease | predicted oxidoreductase, Zn-dependent and NAD(P)-binding | predicted hydrogenase, 4Fe-4S ferredoxin-type component | predicted acetyltransferase | DNA replication/recombination/repair protein | predicted pyruvate formate lyase activating enzyme | hypothetical protein, Qin prophage |
| 89 | 74 | 328 | 0.724 | 0.555 |  |  | 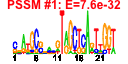 | 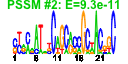 |  | b3570 b3816 b3978 b0148 b0672 b3938 b3396 b2961 b4177 b4246 b2669 b3979 b3976 b2960 b2959 b2962 b3252 b3865 b4251 b4295 b4391 b3002 b4401 b0536 b3796 b3022 b1910 b3472 b4125 b0160 b4232 b4312 b4443 b2864 b3438 b3414 b3797 b4328 b4294 b4369 b4270 b1909 b0147 b4129 b3939 b0192 b4108 b2189 b3799 b4440 b3658 b2695 b4392 b2617 b4448 b3273 b0244 b4393 b3761 b3977 b1665 b1666 b0057 b0163 b0191 b4406 b0190 b0327 b3021 b3068 b3107 b3439 b3820 b4243 | bax corA glyT hrpB leuW metJ mrcA mutY purA pyrL stpA thrT thrU trmI yggL yggX csrD yihA yjgJ yjhU yjjK yqhA arcA argU argX mqsR cysU dcrB dcuS dgt fbp fimB glyU gntR gntY hisR iadA insA leuP leuX leuZ ligT lysU metB nlpE yjdM proL proM serV slt smpA thrV thrW trpR trpT tyrU valV valW yaeH yaeJ yaeP yaeQ yahM ygiT mug yhaL yhhW yigI yjgF | conserved protein | magnesium/nickel/cobalt transporter | tRNA-Gly | predicted ATP-dependent helicase | tRNA-Leu | DNA-binding transcriptional repressor, S-adenosylmethionine-binding | fused penicillin-binding protein 1a: murein transglycosylase/murein transpeptidase | adenine DNA glycosylase | adenylosuccinate synthetase | pyrBI operon leader peptide | DNA binding protein, nucleoid-associated | tRNA-Thr | tRNA (m7G46) methyltransferase, SAM-dependent | predicted protein | protein that protects iron-sulfur proteins against oxidative damage | conserved inner membrane protein | GTP-binding protein | predicted transcriptional regulator | KpLE2 phage-like element; predicted DNA-binding transcriptional regulator | fused predicted transporter subunits of ABC superfamily: ATP-binding components | DNA-binding response regulator in two-component regulatory system with ArcB or CpxA | tRNA-Arg | predicted cyanide hydratase | sulfate/thiosulfate transporter subunit | periplasmic protein | sensory histidine kinase in two-component regulatory system with DcuR, regulator of anaerobic fumarate respiration | deoxyguanosine triphosphate triphosphohydrolase | fructose-1,6-bisphosphatase I | tyrosine recombinase/inversion of on/off regulator of fimA | DNA-binding transcriptional repressor | predicted gluconate transport associated protein | tRNA-His | isoaspartyl dipeptidase | KpLE2 phage-like element; IS1 repressor protein InsA | 2'-5' RNA ligase | lysine tRNA synthetase, inducible | cystathionine gamma-synthase, PLP-dependent | lipoprotein involved with copper homeostasis and adhesion | tRNA-Pro | tRNA-Ser | lytic murein transglycosylase, soluble | small membrane lipoprotein | DNA-binding transcriptional repressor, tryptophan-binding | tRNA-Trp | tRNA-Tyr | tRNA-Val | predicted DNA-binding transcriptional regulator | G/U mismatch-specific DNA glycosylase | ketoacid-binding protein |
| 40 | 41 | 343 | 0.728 | 0.545 |  |  | 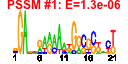 | 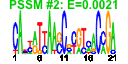 |  | b0646 b2370 b0261 b0708 b2938 b0993 b2046 b2867 b2868 b0117 b4580 b0404 b0412 b0445 b0690 b0876 b2015 b2120 b2338 b2371 b2636 b2637 b2999 b3232 b2940 b3102 b3446 b4223 b0446 b0400 b2819 b0397 b0379 b0456 b0443 b2119 b2835 b0295 b4578 b2466 b2473 | djlB evgS mmuM phr speA torS wzxC xdhB xdhC yacH yaiT acpH yajI ybaE ybjD yeeY yehM yfdE yfjS yfjT yhcM yqgC yqjG yrhB cof phoR recD sbcC yaiY ybaA ybaW yehL lplT ykgL yncK ypfG ypfH | predicted chaperone | hybrid sensory histidine kinase in two-component regulatory system with EvgA | CP4-6 prophage; S-methylmethionine:homocysteine methyltransferase | deoxyribodipyrimidine photolyase, FAD-binding | biosynthetic arginine decarboxylase, PLP-binding | hybrid sensory histidine kinase in two-component regulatory system with TorR | colanic acid exporter | xanthine dehydrogenase, FAD-binding subunit | xanthine dehydrogenase, Fe-S binding subunit | predicted protein | conserved protein | predicted lipoprotein | predicted transporter subunit: periplasmic-binding component of ABC superfamily | conserved protein with nucleoside triphosphate hydrolase domain | predicted DNA-binding transcriptional regulator | predicted CoA-transferase, NAD(P)-binding | CP4-57 prophage; predicted protein | predicted S-transferase | thiamin pyrimidine pyrophosphate hydrolase | sensory histidine kinase in two-component regulatory system with PhoB | exonuclease V (RecBCD complex), alpha chain | exonuclease, dsDNA, ATP-dependent | predicted inner membrane protein | predicted transporter subunit: ATP-binding component of ABC superfamily | predicted hydrolase |
| 11 | 85 | 329 | 0.732 | 0.549 |  |  | 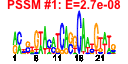 | 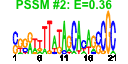 |  | b1623 b0606 b0469 b4409 b0526 b2314 b2511 b2525 b0028 b2321 b0680 b0238 b0477 b2231 b2526 b0026 b0029 b0628 b1117 b0524 b1054 b0027 b1531 b1530 b1047 b3930 b2262 b2261 b2264 b2260 b0661 b2210 b2276 b0741 b2523 b0525 b0441 b0950 b0951 b0194 b0393 b4420 b0058 b1269 b1086 b0610 b1764 b1479 b2522 b0740 b1634 b2232 b0195 b0658 b0742 b0780 b0892 b0905 b0917 b1273 b1626 b1625 b1853 b1840 b1915 b2014 b1983 b2124 b2125 b2126 b2263 b2431 b2524 b2593 b0458 b1841 b2214 b2646 b2791 b2792 b2527 b2528 b2531 b2530 b2529 | add ahpF apt blr cysS dedD der fdx fkpB flk glnS gpt gsk gyrA hscA ileS ispH lipA lolD lpxH lpxL lspA marA marR mdoC menA menB menC menD menE miaB mqo nuoN pal pepB ppiB ppiD pqiA pqiB proS rdgC rluA rluB rluC rnk selD maeA sseB tolB tppB ubiG yaeB ybeX ybgF ybhK rarA ycaO ycaR yciN ydgK cnu yebK yebZ yecF yeeF yeeN yehS yehT yehU yfbB yfeX iscX yfiH ylaC yobA apbE ypjF truC yqcC hscB iscA iscR iscS iscU | adenosine deaminase | alkyl hydroperoxide reductase, F52a subunit, FAD/NAD(P)-binding | adenine phosphoribosyltransferase | beta-lactam resistance membrane protein | cysteinyl-tRNA synthetase | conserved protein | predicted GTP-binding protein | [2Fe-2S] ferredoxin | FKBP-type peptidyl-prolyl cis-trans isomerase (rotamase) | predicted flagella assembly protein | glutamyl-tRNA synthetase | guanine-hypoxanthine phosphoribosyltransferase | inosine/guanosine kinase | DNA gyrase (type II topoisomerase), subunit A | DnaK-like molecular chaperone specific for IscU | isoleucyl-tRNA synthetase | 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase, 4Fe-4S protein | lipoate synthase | outer membrane-specific lipoprotein transporter subunit | UDP-2,3-diacylglucosamine pyrophosphatase | lauryl-acyl carrier protein (ACP)-dependent acyltransferase | prolipoprotein signal peptidase (signal peptidase II) | DNA-binding transcriptional dual activator of multiple antibiotic resistance | DNA-binding transcriptional repressor of multiple antibiotic resistance | membrane protein required for modification of periplasmic glucan | 1,4-dihydroxy-2-naphthoate octaprenyltransferase | dihydroxynaphthoic acid synthetase | o-succinylbenzoyl-CoA synthase | bifunctional 2-oxoglutarate decarboxylase/ SHCHC synthase | o-succinylbenzoate-CoA ligase | isopentenyl-adenosine A37 tRNA methylthiolase | malate dehydrogenase, FAD/NAD(P)-binding domain | NADH:ubiquinone oxidoreductase, membrane subunit N | peptidoglycan-associated outer membrane lipoprotein | aminopeptidase B | peptidyl-prolyl cis-trans isomerase B (rotamase B) | peptidyl-prolyl cis-trans isomerase (rotamase D) | paraquat-inducible membrane protein A | paraquat-inducible protein B | prolyl-tRNA synthetase | DNA-binding protein, non-specific | pseudouridine synthase for 23S rRNA (position 746) and tRNAphe(position 32) | 23S rRNA pseudouridylate synthase | regulator of nucleoside diphosphate kinase | selenophosphate synthase | malate dehydrogenase, (decarboxylating, NAD-requiring) (malic enzyme) | rhodanase-like enzyme, sulfur transfer from thiosulfate | periplasmic protein | predicted transporter | bifunctional 3-demethylubiquinone-9 3-methyltransferase/ 2-octaprenyl-6-hydroxy phenol methylase | predicteed ion transport | predicted protein | predicted transferase with NAD(P)-binding Rossmann-fold domain | recombination protein | conserved inner membrane protein | predicted regulator | predicted DNA-binding transcriptional regulator | predicted inner membrane protein | predicted amino-acid transporter | predicted response regulator in two-component system withYehU | predicted sensory kinase in two-component system with YehT | predicted peptidase | predicted thiamine biosynthesis lipoprotein | CP4-57 prophage; toxin of the YpjF-YfjZ toxin-antitoxin system | tRNA pseudouridine synthase | DnaJ-like molecular chaperone specific for IscU | FeS cluster assembly protein | DNA-binding transcriptional repressor | cysteine desulfurase (tRNA sulfurtransferase), PLP-dependent | scaffold protein |
| 4 | 58 | 340 | 0.749 | 0.647 |  |  |  | 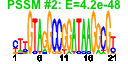 |  | b2048 b3543 b3542 b3541 b2451 b3081 b3868 b3869 b0514 b2787 b2789 b2788 b2489 b4435 b0698 b0697 b0696 b0695 b0694 b4513 b0345 b0403 b3600 b3601 b2711 b1020 b2819 b3902 b2144 b3917 b0188 b3244 b4324 b2044 b0228 b0323 b0689 b0808 b1470 b1509 b1573 b2128 b2129 b2130 b2131 b2145 b2812 b2958 b3082 b3083 b3383 b3901 b3937 b1973 b3070 b3071 b2712 b2322 | cpsG dppB dppC dppD eutA fadH glnG glnL glxK gudD gudP gudX hyfI kdpA kdpB kdpC kdpD kdpE kdpF lacI malZ mtlD mtlR norW phoH recD rhaD sanA sbp tilS tldD uxuR wcaL yafM yahI ybfP ybiO yddJ ydeU rzpQ yehW yehX yehY osmF yeiS ygdL yggN ygjM ygjN yhfZ rhaM yiiX yodA yqjH yqjI hypF yfcJ | phosphomannomutase | dipeptide transporter | reactivating factor for ethanolamine ammonia lyase | 2,4-dienoyl-CoA reductase, NADH and FMN-linked | fused DNA-binding response regulator in two-component regulatory system with GlnL: response regulator/sigma54 interaction protein | sensory histidine kinase in two-component regulatory system with GlnG | glycerate kinase II | (D)-glucarate dehydratase 1 | predicted D-glucarate transporter | predicted glucarate dehydratase | hydrogenase 4, Fe-S subunit | potassium translocating ATPase, subunit A | potassium translocating ATPase, subunit B | potassium translocating ATPase, subunit C | fused sensory histidine kinase in two-component regulatory system with KdpE: signal sensing protein | DNA-binding response regulator in two-component regulatory system with KdpD | potassium ion accessory transporter subunit | DNA-binding transcriptional repressor | maltodextrin glucosidase | mannitol-1-phosphate dehydrogenase, NAD(P)-binding | DNA-binding repressor | NADH:flavorubredoxin oxidoreductase | conserved protein with nucleoside triphosphate hydrolase domain | exonuclease V (RecBCD complex), alpha chain | rhamnulose-1-phosphate aldolase | predicted protein | sulfate transporter subunit | tRNA(Ile)-lysidine synthetase | predicted peptidase | predicted glycosyl transferase | conserved protein | predicted carbamate kinase-like protein | predicted mechanosensitive channel | predicted transporter subunit: membrane component of ABC superfamily | predicted transporter subunit: ATP-binding component of ABC superfamily | predicted transporter subunit: periplasmic-binding component of ABC superfamily | predicted inner membrane protein | predicted DNA-binding transcriptional regulator | L-rhamnose mutarotase | predicted peptidoglycan peptidase | conserved metal-binding protein | predicted siderophore interacting protein | predicted transcriptional regulator | carbamoyl phosphate phosphatase and maturation protein for [NiFe] hydrogenases | predicted transporter |
| 134 | 79 | 324 | 0.765 | 0.490 |  |  | 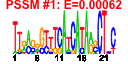 | 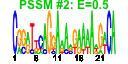 |  | b1176 b1175 b2564 b2293 b2563 b0376 b0863 b0861 b0864 b0862 b1992 b1993 b1538 b1594 b0604 b2267 b1651 b0212 b0962 b1103 b1250 b1827 b2262 b1114 b1174 b1326 b1068 b1740 b1107 b2193 b2418 b1130 b1101 b1854 b1066 b1427 b1234 b1235 b1323 b2509 b0213 b0577 b0711 b0712 b0925 b0956 b0927 b0965 b1033 b1067 b1128 b1100 b1104 b1105 b1106 b1108 b1162 b1539 b1540 b1614 b1647 b1664 b1667 b1765 b1958 b2016 b2177 b2291 b2352 b2379 b2419 b2494 b2495 b2493 b1585 b1586 b2136 b2474 b2475 | minC minD pdxJ yfbT acpS ampH artI artM artP artQ cobS cobU dcp dgsA dsbG elaA gloA gloB helD hinT kch kdgR menB mfd minE mpaA yceM nadE nagZ narP pdxK phoP ptsG pykA rimJ rimL rssA rssB tyrR xseA yafS ybdG ybgJ ybgK ycbB ycbG ycbL yccU ghrA yceH ycfD ycfH ycfL ycfM thiK ycfP ycgE ydfG ydfH ydgA ydhF ydhQ ydhR ydjA yedI yeeZ yejA yfbR yfdI yfdZ yfeK yfgC yfgD yfgO ynfC ynfD yohD ypfI ypfJ | cell division inhibitor | membrane ATPase of the MinC-MinD-MinE system | pyridoxine 5'-phosphate synthase | predicted hydrolase or phosphatase | holo-[acyl-carrier-protein] synthase 1 | beta-lactamase/D-alanine carboxypeptidase | arginine transporter subunit | cobalamin 5'-phosphate synthase | bifunctional cobinamide kinase/ cobinamide phosphate guanylyltransferase | dipeptidyl carboxypeptidase II | DNA-binding transcriptional repressor | periplasmic disulfide isomerase/thiol-disulphide oxidase | predicted acyltransferase with acyl-CoA N-acyltransferase domain | glyoxalase I, Ni-dependent | predicted hydroxyacylglutathione hydrolase | DNA helicase IV | purine nucleoside phosphoramidase | voltage-gated potassium channel | predicted DNA-binding transcriptional regulator | dihydroxynaphthoic acid synthetase | transcription-repair coupling factor | cell division topological specificity factor | murein peptide amidase A | predicted oxidoreductase with NAD(P)-binding Rossmann-fold domain | NAD synthetase, NH3/glutamine-dependent | beta N-acetyl-glucosaminidase | DNA-binding response regulator in two-component regulatory system with NarQ or NarX | pyridoxal-pyridoxamine kinase/hydroxymethylpyrimidine kinase | DNA-binding response regulator in two-component regulatory system with PhoQ | fused glucose-specific PTS enzymes: IIB component/IIC component | pyruvate kinase II | ribosomal-protein-S5-alanine N-acetyltransferase | ribosomal-protein-L7/L12-serine acetyltransferase | conserved protein | response regulator of RpoS | DNA-binding transcriptional dual regulator, tyrosine-binding | exonuclease VII, large subunit | predicted S-adenosyl-L-methionine-dependent methyltransferase | predicted mechanosensitive channel | predicted enzyme subunit | predicted carboxypeptidase | predicted metal-binding enzyme | predicted CoA-binding protein with NAD(P)-binding Rossmann-fold domain | 2-ketoacid reductase | predicted metallodependent hydrolase | predicted protein | predicted outer membrane lipoprotein | thiamin kinase | L-allo-threonine dehydrogenase, NAD(P)-binding | predicted oxidoreductase | conserved inner membrane protein | predicted epimerase, with NAD(P)-binding Rossmann-fold domain | predicted oligopeptide transporter subunit | deoxyribonucleoside 5'-monophosphatase | CPS-53 (KpLE1) prophage; predicted inner membrane protein | prediected aminotransferase, PLP-dependent | predicted peptidase | predicted inner membrane protein | predicted hydrolase |
| 36 | 49 | 339 | 0.767 | 0.593 |  |  | 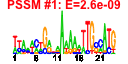 | 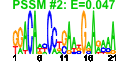 |  | b2182 b2456 b0619 b0620 b2268 b0543 b2453 b2454 b1379 b0537 b2349 b0465 b0657 b4570 b1304 b1306 b1307 b1303 b2183 b0293 b0443 b0510 b0659 b0660 b0709 b1161 b1505 b1549 b1877 b2102 b2184 b2244 b2352 b4578 b4533 b1796 b4536 b2462 b1859 b2261 b2260 b1907 b0899 b1381 b1573 b1929 b1930 b3267 b2107 | bcr eutN dpiB dpiA rbn emrE eutG eutJ hslJ intD intS kefA lnt lomR pspA pspC pspD pspF rsuA matB ybaW ybeY ybeZ ybgH ycgX ydeT ydfO yecT yegX yejH yfaD yfdI yncK ynfO yoaG yobH eutS znuB menC menE tyrP ycaM ydbH rzpQ yedE yedF yhdV yohN | bicyclomycin/multidrug efflux system | predicted carboxysome structural protein, ethanolamine utilization protein | sensory histidine kinase in two-component regulatory system with citB | DNA-binding response regulator in two-component regulatory system with citA | binuclear zinc phosphodiesterase | DLP12 prophage; multidrug resistance protein | predicted alcohol dehydrogenase in ethanolamine utilization | predicted chaperonin, ethanolamine utilization protein | heat-inducible protein | DLP12 prophage; predicted integrase | CPS-53 (KpLE1) prophage; predicted prophage CPS-53 integrase | fused conserved protein | apolipoprotein N-acyltransferase | regulatory protein for phage-shock-protein operon | DNA-binding transcriptional activator | peripheral inner membrane phage-shock protein | 16S rRNA pseudouridylate 516 synthase | conserved protein | predicted protein with nucleoside triphosphate hydrolase domain | predicted transporter | predicted protein | Qin prophage; predicted protein | predicted hydrolase | predicted ATP-dependet helicase | CPS-53 (KpLE1) prophage; predicted inner membrane protein | hypothetical protein, Qin prophage | predicted carboxysome structural protein with predicted role in ethanol utilization | zinc transporter subunit: membrane component of ABC superfamily | o-succinylbenzoyl-CoA synthase | o-succinylbenzoate-CoA ligase | tyrosine transporter | predicted inner membrane protein | predicted outer membrane protein |
| 92 | 107 | 335 | 0.777 | 0.547 |  |  | 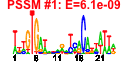 | 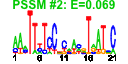 |  | b2219 b4152 b3526 b3115 b3496 b3873 b4299 b4357 b1002 b4055 b1901 b4139 b4213 b2535 b4138 b4123 b3093 b0048 b4154 b4153 b4151 b3946 b2800 b2802 b2799 b2801 b2805 b4122 b4140 b2151 b2996 b2995 b2994 b2993 b2992 b2991 b2990 b2997 b4036 b3605 b3603 b3604 b4034 b4033 b4032 b4035 b3225 b3223 b3224 b4238 b4237 b2964 b4021 b3753 b0030 b4089 b4438 b3118 b3117 b3116 b3114 b3113 b4471 b3708 b3709 b3707 b2426 b3092 b4322 b4323 b0380 b0791 b1426 b2147 b2146 b2251 b2399 b2428 b2597 b2869 b2877 b2936 b3001 b3020 b3087 b3156 b3221 b3872 b4188 b4252 b4279 b4280 b4297 b4298 b4306 b4353 b4354 b0306 b0307 b0308 b1137 b1138 b2875 b2876 b2844 b4216 b4227 | atoS frdC kdgK tdcD yhiP yihM yjhI yjjM agp aphA araF aspA cpdB csiE dcuA dcuB exuT folA frdA frdB frdD fsaB fucA fucI fucO fucP fucR fumB fxsA galS hybA hybB hybC hybD hybE hybF hybG hybO lamB lldD lldP lldR malE malF malG malK nanA nanE nanT nrdD nrdG nupG pepE rbsR rihC rpiR tdcA tdcB tdcC tdcE tdcF tdcG tnaA tnaB tnaC ucpA uxaC uxuA uxuB yaiZ ybhQ ydcH yeiA yeiT nudI yfeD murQ raiA ygeV ygfJ yggG yghZ ygiS ygjR yhbS yhcH yihL yjfN yjgK yjhB yjhC yjhG yjhH yjhP yjiX yjiY ykgE ykgF ykgG ymfD ymfE yqeB yqeC yqeF ytfJ ytfQ | sensory histidine kinase in two-component regulatory system with AtoC | fumarate reductase (anaerobic), membrane anchor subunit | ketodeoxygluconokinase | propionate kinase/acetate kinase C, anaerobic | predicted transporter | predicted sugar phosphate isomerase | KpLE2 phage-like element; predicted DNA-binding transcriptional regulator | predicted DNA-binding transcriptional regulator | glucose-1-phosphatase/inositol phosphatase | acid phosphatase/phosphotransferase, class B, non-specific | L-arabinose transporter subunit | aspartate ammonia-lyase | 2':3'-cyclic-nucleotide 2'-phosphodiesterase | stationary phase inducible protein | C4-dicarboxylate antiporter | hexuronate transporter | dihydrofolate reductase | fumarate reductase (anaerobic) catalytic and NAD/flavoprotein subunit | fumarate reductase (anaerobic), Fe-S subunit | fructose-6-phosphate aldolase 2 | L-fuculose-1-phosphate aldolase | L-fucose isomerase | L-1,2-propanediol oxidoreductase | L-fucose transporter | DNA-binding transcriptional activator | anaerobic class I fumarate hydratase (fumarase B) | inner membrane protein | DNA-binding transcriptional repressor | hydrogenase 2 4Fe-4S ferredoxin-type component | predicted hydrogenase 2 cytochrome b type component | hydrogenase 2, large subunit | predicted maturation element for hydrogenase 2 | hydrogenase 2-specific chaperone | protein involved with the maturation of hydrogenases 1 and 2 | hydrogenase 2 accessory protein | hydrogenase 2, small subunit | maltose outer membrane porin (maltoporin) | L-lactate dehydrogenase, FMN-linked | L-lactate permease | maltose transporter subunit | fused maltose transport subunit, ATP-binding component of ABC superfamily/regulatory protein | N-acetylneuraminate lyase | predicted N-acetylmannosamine-6-P epimerase | sialic acid transporter | anaerobic ribonucleoside-triphosphate reductase | anaerobic ribonucleotide reductase activating protein | nucleoside transporter | (alpha)-aspartyl dipeptidase | DNA-binding transcriptional repressor of ribose metabolism | ribonucleoside hydrolase 3 | catabolic threonine dehydratase, PLP-dependent | L-threonine/L-serine transporter | pyruvate formate-lyase 4/2-ketobutyrate formate-lyase | predicted L-PSP (mRNA) endoribonuclease | L-serine dehydratase 3 | tryptophanase/L-cysteine desulfhydrase, PLP-dependent | tryptophan transporter of low affinity | tryptophanase leader peptide | predicted oxidoredutase, sulfate metabolism protein | uronate isomerase | mannonate hydrolase | D-mannonate oxidoreductase, NAD-binding | predicted inner membrane protein | predicted protein | predicted oxidoreductase | predicted NUDIX hydrolase | predicted PTS component | cold shock protein associated with 30S ribosomal subunit | conserved protein | predicted peptidase | aldo-keto reductase | predicted transporter subunit: periplasmic-binding component of ABC superfamily | predicted NAD(P)-binding dehydrogenase | predicted acyltransferase with acyl-CoA N-acyltransferase domain | KpLE2 phage-like element; predicted transporter | KpLE2 phage-like element; predicted oxidoreductase | KpLE2 phage-like element; predicted dehydratase | KpLE2 phage-like element; predicted lyase/synthase | KpLE2 phage-like element; predicted methyltransferase | predicted amino acid dehydrogenase with NAD(P)-binding domain and ferridoxin-like domain | e14 prophage; predicted SAM-dependent methyltransferase | e14 prophage; predicted inner membrane protein | conserved protein with NAD(P)-binding Rossman fold | predicted acyltransferase | predicted transcriptional regulator | predicted sugar transporter subunit: periplasmic-binding component of ABC superfamily |
| 38 | 87 | 336 | 0.805 | 0.563 |  |  | 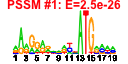 | 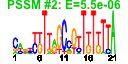 |  | b3133 b3588 b4087 b4479 b3077 b3075 b3093 b3845 b3846 b3950 b3949 b3953 b3946 b2800 b2802 b2801 b3128 b3124 b3126 b3127 b3125 b4267 b4119 b3223 b4238 b4237 b3951 b3947 b3421 b4565 b3118 b3117 b3116 b3114 b3113 b4471 b3709 b4196 b3092 b4322 b4323 b2866 b2341 b2342 b2869 b2877 b2981 b3106 b3154 b3155 b3573 b3674 b3676 b3677 b4253 b4311 b4326 b4335 b4379 b4506 b2875 b2876 b3152 b4003 b3665 b3135 b3088 b3371 b4474 b3945 b4321 b2787 b2788 b3132 b4475 b4195 b4197 b3091 b2632 b3074 b3110 b3664 b3675 b0296 b2966 b4209 b4004 | agaV aldB alsA dgoR ebgC ebgR exuT fadA fadB frwB frwC frwD fsaB fucA fucI fucP garD garK garL garP garR idnD melA nanE nrdD nrdG pflD ptsA rtcB sgcB tdcA tdcB tdcC tdcE tdcF tdcG tnaB ulaD uxaC uxuA uxuB xdhA fadJ fadI ygeV ygfJ yghO yhaK yhbP yhbQ ysaA yidF yidH yidI yjgL nanC yjiD yjiM yjjW ykgO yqeB yqeC yraR zraS ade agaA alx frlB frlC gldA gntP gudD gudX kbaZ rtcA ulaC ulaE uxaA yfjP ygjH yhaO yicO yidG ykgM yqgA ytfE zraR | N-acetylgalactosamine-specific enzyme IIB component of PTS | aldehyde dehydrogenase B | fused D-allose transporter subunits of ABC superfamily: ATP-binding components | predicted DNA-binding transcriptional regulator | cryptic beta-D-galactosidase, beta subunit | DNA-binding transcriptional repressor | hexuronate transporter | 3-ketoacyl-CoA thiolase (thiolase I) | fused 3-hydroxybutyryl-CoA epimerase/delta(3)-cis-delta(2)-trans-enoyl-CoA isomerase/enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase | predicted enzyme IIB component of PTS | predicted enzyme IIC component of PTS | fructose-6-phosphate aldolase 2 | L-fuculose-1-phosphate aldolase | L-fucose isomerase | L-fucose transporter | (D)-galactarate dehydrogenase | glycerate kinase I | alpha-dehydro-beta-deoxy-D-glucarate aldolase | predicted (D)-galactarate transporter | tartronate semialdehyde reductase | L-idonate 5-dehydrogenase, NAD-binding | alpha-galactosidase, NAD(P)-binding | predicted N-acetylmannosamine-6-P epimerase | anaerobic ribonucleoside-triphosphate reductase | anaerobic ribonucleotide reductase activating protein | predicted formate acetyltransferase 2 (pyruvate formate lyase II) | fused predicted PTS enzymes: Hpr component/enzyme I component/enzyme IIA component | conserved protein | DNA-binding transcriptional activator | catabolic threonine dehydratase, PLP-dependent | L-threonine/L-serine transporter | pyruvate formate-lyase 4/2-ketobutyrate formate-lyase | predicted L-PSP (mRNA) endoribonuclease | L-serine dehydratase 3 | tryptophan transporter of low affinity | 3-keto-L-gulonate 6-phosphate decarboxylase | uronate isomerase | mannonate hydrolase | D-mannonate oxidoreductase, NAD-binding | xanthine dehydrogenase, molybdenum binding subunit | fused enoyl-CoA hydratase and epimerase and isomerase/3-hydroxyacyl-CoA dehydrogenase | beta-ketoacyl-CoA thiolase, anaerobic, subunit | predicted pirin-related protein | predicted endonuclease | predicted hydrogenase, 4Fe-4S ferredoxin-type component | conserved inner membrane protein | predicted inner membrane protein | predicted protein | N-acetylnuraminic acid outer membrane channel protein | DNA replication/recombination/repair protein | predicted 2-hydroxyglutaryl-CoA dehydratase | predicted pyruvate formate lyase activating enzyme | rpmJ (L36) paralog | conserved protein with NAD(P)-binding Rossman fold | predicted nucleoside-diphosphate-sugar epimerase | sensory histidine kinase in two-component regulatory system with ZraR | cryptic adenine deaminase | predicted inner membrane protein, part of terminus | fructoselysine-6-P-deglycase | predicted isomerase | glycerol dehydrogenase, NAD | fructuronate transporter | (D)-glucarate dehydratase 1 | predicted glucarate dehydratase | tagatose 6-phosphate aldolase 1, kbaZ subunit | RNA 3'-terminal phosphate cyclase | L-ascorbate-specific enzyme IIA component of PTS | L-xylulose 5-phosphate 3-epimerase | altronate hydrolase | CP4-57 prophage; predicted GTP-binding protein | predicted transporter | predicted xanthine/uracil permase | predicted regulator of cell morphogenesis and cell wall metabolism | fused DNA-binding response regulator in two-component regulatory system with ZraS: response regulator/sigma54 interaction protein |
| 105 | 80 | 337 | 0.864 | 0.587 |  |  |  |  |  | b0557 b0126 b2267 b0852 b2582 b0127 b0128 b1055 b1106 b1572 b1649 b1706 b1825 b1968 b2581 b1826 b2435 b2817 b0433 b3057 b0158 b1216 b2472 b0207 b2268 b3408 b3409 b0588 b0590 b0589 b4367 b0683 b0124 b0755 b2436 b0029 b2587 b1599 b2392 b0817 b2210 b0086 b2675 b2674 b2134 b0467 b1305 b0214 b0739 b0737 b0738 b1252 b2497 b0006 b0195 b0315 b0387 b0427 b0466 b0468 b0591 b0580 b4511 b0736 b1445 b1571 b1643 b1705 b1707 b2432 b2433 b2558 b2807 b3410 b4567 b0991 b1451 b1452 b4536 b2211 | borD can elaA rimK trxC yadG yadH yceA thiK ydfB ydhM ydiU yebO yedV yfiF mgrB amiA amiC ampG bacA btuF chaA dapE dkgB rbn feoA feoB fepC fepD fepG fhuF fur gcd gpmA hemF ispH kgtP mdtI mntH mntR mqo murF nrdE nrdI pbpG priC pspB rnhA tolA tolQ tolR tonB uraA yaaA yaeB yahA yaiI yajR ybaM ybaN entS ybdJ ybdZ ybgC ydcX ydfA ydhI ydiE ydiV yfeY yfeZ yfhD ygdD feoC yjjZ ymcE yncD yncE yobH yojI | DLP12 prophage; predicted lipoprotein | carbonic anhydrase | predicted acyltransferase with acyl-CoA N-acyltransferase domain | ribosomal protein S6 modification protein | thioredoxin 2 | predicted transporter subunit: ATP-binding component of ABC superfamily | predicted transporter subunit: membrane component of ABC superfamily | conserved protein | thiamin kinase | Qin prophage; predicted protein | predicted DNA-binding transcriptional regulator | predicted protein | predicted sensory kinase in two-component regulatory system with YedW | predicted methyltransferase | N-acetylmuramoyl-l-alanine amidase I | N-acetylmuramoyl-L-alanine amidase | muropeptide transporter | undecaprenyl pyrophosphate phosphatase | vitamin B12 transporter subunit: periplasmic-binding component of ABC superfamily | calcium/sodium:proton antiporter | N-succinyl-diaminopimelate deacylase | 2,5-diketo-D-gluconate reductase B | binuclear zinc phosphodiesterase | ferrous iron transporter, protein A | fused ferrous iron transporter, protein B: GTP-binding protein/membrane protein | iron-enterobactin transporter subunit | ferric iron reductase involved in ferric hydroximate transport | DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport | glucose dehydrogenase | phosphoglyceromutase 1 | coproporphyrinogen III oxidase | 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase, 4Fe-4S protein | alpha-ketoglutarate transporter | multidrug efflux system transporter | manganese/divalent cation transporter | DNA-binding transcriptional regulator of mntH | malate dehydrogenase, FAD/NAD(P)-binding domain | UDP-N-acetylmuramoyl-tripeptide:D-alanyl-D- alanine ligase | ribonucleoside-diphosphate reductase 2, alpha subunit | protein that stimulates ribonucleotide reduction | D-alanyl-D-alanine endopeptidase | primosomal replication protein N'' | DNA-binding transcriptional regulator of psp operon | ribonuclease HI, degrades RNA of DNA-RNA hybrids | membrane anchored protein in TolA-TolQ-TolR complex | membrane spanning protein in TolA-TolQ-TolR complex | membrane spanning protein in TonB-ExbB-ExbD complex | uracil transporter | predicted transporter | conserved inner membrane protein | predicted inner membrane protein | predicted acyl-CoA thioesterase | predicted transglycosylase | cold shock gene | predicted iron outer membrane transporter | fused predicted multidrug transport subunits of ABC superfamily: membrane component/ATP-binding component |
| 3 | 61 | 338 | 0.872 | 0.500 |  |  |  |  |  | b3240 b0039 b1575 b1569 b1574 b0554 b2731 b0825 b0479 b2799 b4415 b2712 b0255 b3132 b2839 b0230 b0018 b2429 b4095 b1158 b0402 b0700 b4436 b4437 b0068 b0066 b1666 b0024 b0065 b0267 b0268 b0272 b0442 b4508 b1013 b1154 b1239 b1228 b1355 b1568 b1790 b4496 b2119 b4502 b4544 b2322 b2337 b2356 b2732 b2776 b2766 b2767 b2768 b2872 b2902 b2930 b2950 b3489 b0253 b2473 b2545 | aaeB caiA dicB dicC essD fhlA fsaA fsr fucO hokE hypF kbaZ lysR lafU mokC murP phnM pinE proY rhsC tbpA thiQ valW yaaY yabI yagA yagE yagI ybaV ybcD rutR ycfK ychS ydaG ydfX yeaM yedS yehL yeiW yfbW yfcJ yfdM ygbA ygcE ygcN ygcO ygcP ygeY ygfF yggF yggR ykfA ypfH yphC | p-hydroxybenzoic acid efflux system component | crotonobetaine reductase subunit II, FAD-binding | Qin prophage; cell division inhibition protein | Qin prophage; DNA-binding transcriptional regulator for DicB | DLP12 prophage; predicted phage lysis protein | DNA-binding transcriptional activator | fructose-6-phosphate aldolase 1 | predicted fosmidomycin efflux system | L-1,2-propanediol oxidoreductase | toxic polypeptide, small | carbamoyl phosphate phosphatase and maturation protein for [NiFe] hydrogenases | tagatose 6-phosphate aldolase 1, kbaZ subunit | DNA-binding transcriptional dual regulator | regulatory protein for HokC, overlaps CDS of hokC | fused predicted PTS enzymes: IIB component/IIC component | carbon-phosphorus lyase complex subunit | e14 prophage; site-specific DNA recombinase | predicted cryptic proline transporter | rhsC element core protein RshC | thiamin transporter subunit | tRNA-Val | predicted protein | conserved inner membrane protein | CP4-6 prophage; predicted DNA-binding transcriptional regulator | CP4-6 prophage; predicted lyase/synthase | conserved protein | predicted DNA-binding transcriptional regulator | e14 prophage; predicted protein | Rac prophage; predicted protein | Qin prophage; predicted protein | predicted transporter subunit: ATP-binding component of ABC superfamily | predicted transporter | predicted kinase | predicted oxidoreductase with FAD/NAD(P)-binding domain | predicted 4Fe-4S cluster-containing protein | predicted anti-terminator regulatory protein | predicted peptidase | predicted NAD(P)-binding oxidoreductase with NAD(P)-binding Rossmann-fold domain | predicted hexoseP phosphatase | CP4-6 prophage; predicted GTP-binding protein | predicted hydrolase | predicted oxidoreductase, Zn-dependent and NAD(P)-binding |
| 29 | 74 | 342 | 0.883 | 0.565 |  |  |  |  |  | b0110 b0111 b0564 b0446 b0338 b0340 b0339 b0341 b0166 b0207 b0221 b0142 b2167 b2168 b2804 b0200 b1550 b4328 b3069 b0047 b0046 b0075 b1677 b0889 b0109 b0714 b2215 b1243 b1244 b1247 b2599 b2598 b1299 b0497 b0070 b0627 b0129 b0210 b0266 b0277 b0287 b0382 b0454 b0488 b0489 b0491 b0579 b0710 b0762 b0798 b0800 b1205 b4492 b2008 b2181 b2291 b3105 b3253 b4330 b0502 b1053 b0260 b0069 b2340 b2965 b0068 b0130 b0546 b0792 b0793 b4064 b4329 b2141 b2142 | ampD ampE appY cof cynR cynS cynT cynX dapD dkgB fadE folK fruA fruK fucU gmhB gnsB iadA ileX kefC kefF leuL lpp lrp nadC nei ompC oppA oppB oppF pheA pheL puuR rhsD setA tatE yadI yafE yagB yagK yagU iraP ybaZ ybbJ qmcA ybbM ybdF ybgI ybhT ybiA ybiB ychH ydbA yeeA yejG yfbR yhaJ yhdH yjiH ylbG mdtG mmuP sgrR sixA speC tbpA yadE ybcM ybhR ybhS yjcD yjiG yohJ yohK | N-acetyl-anhydromuranmyl-L-alanine amidase | predicted inner membrane protein | DLP12 prophage; DNA-binding transcriptional activator | thiamin pyrimidine pyrophosphate hydrolase | DNA-binding transcriptional dual regulator | cyanate aminohydrolase | carbonic anhydrase | predicted cyanate transporter | 2,3,4,5-tetrahydropyridine-2-carboxylate N-succinyltransferase | 2,5-diketo-D-gluconate reductase B | acyl coenzyme A dehydrogenase | 2-amino-4-hydroxy-6-hydroxymethyldihyropteridine pyrophosphokinase | fused fructose-specific PTS enzymes: IIBcomponent/IIC components | fructose-1-phosphate kinase | L-fucose mutarotase | D,D-heptose 1,7-bisphosphate phosphatase | Qin prophage; predicted protein | isoaspartyl dipeptidase | tRNA-Ile | potassium:proton antiporter | flavoprotein subunit for the KefC potassium efflux system | leu operon leader peptide | murein lipoprotein | DNA-binding transcriptional dual regulator, leucine-binding | quinolinate phosphoribosyltransferase | endonuclease VIII/ 5-formyluracil/5-hydroxymethyluracil DNA glycosylase | outer membrane porin protein C | oligopeptide transporter subunit | fused chorismate mutase P/prephenate dehydratase | pheA gene leader peptide | DNA-binding transcriptional repressor | rhsD element protein | broad specificity sugar efflux system | TatABCE protein translocation system subunit | predicted PTS Enzyme IIA | predicted S-adenosyl-L-methionine-dependent methyltransferase | CP4-6 prophage; conserved protein | conserved inner membrane protein | predicted protein | predicted methyltransferase | predicted protease, membrane anchored | conserved protein | conserved metal-binding protein | predicted transferase/phosphorylase | deoxyribonucleoside 5'-monophosphatase | predicted DNA-binding transcriptional regulator | predicted oxidoreductase, Zn-dependent and NAD(P)-binding | predicted drug efflux system | CP4-6 prophage; predicted S-methylmethionine transporter | DNA-binding transcriptional regulator | phosphohistidine phosphatase | ornithine decarboxylase, constitutive | thiamin transporter subunit | predicted polysaccharide deacetylase lipoprotein | DLP12 prophage; predicted DNA-binding transcriptional regulator | predicted transporter subunit: membrane component of ABC superfamily | predicted permease |
| 12 | 78 | 329 | 0.889 | 0.533 |  |  |  | 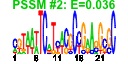 |  | b0885 b0861 b0930 b2027 b0887 b2010 b0840 b2440 b1805 b2042 b1236 b1829 b2442 b1114 b1782 b0761 b1329 b1633 b2134 b1715 b0576 b0399 b1129 b0611 b1627 b1630 b1632 b1631 b2011 b0398 b2340 b1641 b1323 b2028 b2580 b1913 b0444 b0807 b0841 b0868 b4573 b1267 b1266 b1445 b1540 b1722 b1786 b1798 b1928 b2081 b2101 b2438 b2510 b2493 b1145 b1330 b1451 b1824 b2107 b2511 b1054 b1047 b0441 b0950 b0194 b1269 b1086 b1479 b2522 b0780 b0905 b1626 b1625 b2014 b2124 b2125 b2126 b2214 | aat artM asnS cld cydD dacD deoR eutC fadD galF galU htpX intZ mfd mipA modE mppA nth pbpG pheM pheP phoB phoQ rna rsxA rsxD rsxE rsxG sbcB sbcD sixA slyB tyrR ugd ung uvrC queC ybiN ybjG ybjS ychG yciO trpH ydcX ydfH ydiY yeaJ leuE yedD yegQ yegW eutK yfgJ yfgO ymfK ynaI yncD yobF yohN der lpxL mdoC ppiD pqiA proS rluB rluC maeA sseB ybhK ycaO ydgK cnu yeeF yehS yehT yehU apbE | leucyl/phenylalanyl-tRNA-protein transferase | arginine transporter subunit | asparaginyl tRNA synthetase | regulator of length of O-antigen component of lipopolysaccharide chains | fused cysteine transporter subunits of ABC superfamily: membrane component/ATP-binding component | D-alanyl-D-alanine carboxypeptidase (penicillin-binding protein 6b) | DNA-binding transcriptional repressor | ethanolamine ammonia-lyase, small subunit (light chain) | acyl-CoA synthetase (long-chain-fatty-acid--CoA ligase) | predicted subunit with GalU | glucose-1-phosphate uridylyltransferase | predicted endopeptidase | CPZ-55 prophage; predicted integrase | transcription-repair coupling factor | scaffolding protein for murein synthesizing machinery | DNA-binding transcriptional dual regulator | murein tripeptide (L-ala-gamma-D-glutamyl-meso-DAP) transporter subunit | DNA glycosylase and apyrimidinic (AP) lyase (endonuclease III) | D-alanyl-D-alanine endopeptidase | phenylalanyl-tRNA synthetase operon leader peptide | phenylalanine transporter | DNA-binding response regulator in two-component regulatory system with PhoR (or CreC) | sensory histidine kinase in two-compoent regulatory system with PhoP | ribonuclease I | predicted inner membrane subunit | predicted inner membrane oxidoreductase | predicted inner membrane NADH-quinone reductase | predicted oxidoreductase | exonuclease I | exonuclease, dsDNA, ATP-dependent | phosphohistidine phosphatase | outer membrane lipoprotein | DNA-binding transcriptional dual regulator, tyrosine-binding | UDP-glucose 6-dehydrogenase | uracil-DNA-glycosylase | excinuclease UvrABC, endonuclease subunit | predicted aluminum resistance protein | predicted SAM-dependent methyltransferase | undecaprenyl pyrophosphate phosphatase | predicted NAD(P)H-binding oxidoreductase with NAD(P)-binding Rossmann-fold domain | conserved protein | predicted inner membrane protein | predicted DNA-binding transcriptional regulator | predicted diguanylate cyclase | neutral amino-acid efflux system | predicted protein | predicted peptidase | predicted carboxysome structural protein with predicted role in ethanolamine utilization | e14 prophage; repressor protein phage e14 | conserved inner membrane protein | predicted iron outer membrane transporter | predicted GTP-binding protein | lauryl-acyl carrier protein (ACP)-dependent acyltransferase | membrane protein required for modification of periplasmic glucan | peptidyl-prolyl cis-trans isomerase (rotamase D) | paraquat-inducible membrane protein A | prolyl-tRNA synthetase | 23S rRNA pseudouridylate synthase | malate dehydrogenase, (decarboxylating, NAD-requiring) (malic enzyme) | rhodanase-like enzyme, sulfur transfer from thiosulfate | predicted transferase with NAD(P)-binding Rossmann-fold domain | predicted regulator | predicted amino-acid transporter | predicted response regulator in two-component system withYehU | predicted sensory kinase in two-component system with YehT | predicted thiamine biosynthesis lipoprotein |
| 39 | 61 | 327 | 0.928 | 0.541 |  |  | 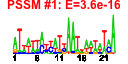 | 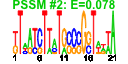 |  | b2366 b1556 b1507 b1508 b1182 b0650 b2727 b1620 b1621 b0569 b1353 b1376 b0498 b0545 b0702 b0704 b0909 b1021 b1022 b1023 b1024 b1025 b1029 b1111 b1537 b1497 b1501 b1503 b1504 b1555 b1674 b1675 b1785 b1895 b2063 b2085 b2083 b2147 b2157 b2158 b2190 b2250 b2305 b2360 b2395 b2558 b0499 b1365 b1810 b1974 b2105 b2106 b0241 b0497 b0787 b1013 b1499 b1502 b1505 b1821 b1796 | dsdA essQ hipA hipB hlyE hscC hypB malI malX nfrB sieB uspF ybbC ybcL ybfB ybfC ycaL pgaD pgaC pgaB pgaA ycdT ycdU ycfQ ydeJ ydeM ydeP ydeR ydeS ydfR ydhY ydhZ yeaI uspC yegH yegR yegZ yeiA yeiE yeiH yejO yfaZ yfcI yfdQ yfeA yfhD ylbH ynaK yoaC yodB rcnR rcnA phoE rhsD ybhM rutR ydeO ydeQ ydeT yebN yoaG | D-serine ammonia-lyase | Qin prophage; predicted S lysis protein | regulator with hipB | DNA-binding transcriptional regulator | hemolysin E | Hsp70 family chaperone Hsc62, binds to RpoD and inhibits transcription | GTP hydrolase involved in nickel liganding into hydrogenases | DNA-binding transcriptional repressor | fused maltose and glucose-specific PTS enzymes: IIB component -! IIC component | bacteriophage N4 receptor, inner membrane subunit | Rac prophage; phage superinfection exclusion protein | stress-induced protein, ATP-binding protein | predicted protein | DLP12 prophage; predicted kinase inhibitor | predicted inner membrane protein | predicted peptidase with chaperone function | predicted glycosyl transferase | predicted enzyme associated with biofilm formation | predicted outer membrane protein | predicted diguanylate cyclase | predicted DNA-binding transcriptional regulator | conserved protein | predicted oxidoreductase | predicted fimbrial-like adhesin protein | Qin prophage; predicted protein | predicted 4Fe-4S ferridoxin-type protein | universal stress protein | fused predicted membrane protein/predicted membrane protein | conserved inner membrane protein | predicted outer membrane porin protein | CPS-53 (KpLE1) prophage; predicted protein | predicted transglycosylase | Rac prophage; conserved protein | predicted cytochrome | membrane protein conferring nickel and cobalt resistance | outer membrane phosphoporin protein E | rhsD element protein | predicted DNA-binding transcriptional acfivator |
| 58 | 54 | 337 | 0.952 | 0.549 |  |  | 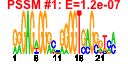 | 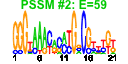 |  | b0433 b2686 b1288 b2803 b0475 b1562 b1370 b0147 b0817 b0149 b3481 b0269 b0270 b0271 b0427 b0528 b0846 b1868 b2467 b0816 b1808 b4546 b2348 b2714 b2182 b0340 b0543 b3174 b0230 b0261 b0018 b0576 b0243 b3545 b4436 b0188 b0268 b0272 b4508 b0771 b0815 b0818 b0868 b1058 b1111 b1242 b1228 b2173 b4502 b2184 b2510 b2902 b3232 b0832 | ampG emrB fabI fucK hemH hokD insH ligT mntR mrcB nikR yagF yagG yagH yajR ybcJ ybjK yecE nudK yliL yoaA ypeB argW ascG bcr cynS emrE leuU lafU mmuM mokC pheP proA proK tilS yagE yagI ybcD ybhJ ybiP ybiR ybjS yceO ycfQ ychE ychS yeiR yeiW yejH yfgJ ygfF yhcM gsiD | muropeptide transporter | multidrug efflux system protein | enoyl-[acyl-carrier-protein] reductase, NADH-dependent | L-fuculokinase | ferrochelatase | Qin prophage; small toxic polypeptide | IS5 transposase and trans-activator | 2'-5' RNA ligase | DNA-binding transcriptional regulator of mntH | fused glycosyl transferase and transpeptidase | DNA-binding transcriptional repressor, Ni-binding | CP4-6 prophage; predicted dehydratase | CP4-6 prophage; predicted sugar transporter | CP4-6 prophage; predicted xylosidase/arabinosidase | predicted transporter | predicted RNA-binding protein | predicted DNA-binding transcriptional regulator | conserved protein | predicted NUDIX hydrolase | predicted protein | conserved protein with nucleoside triphosphate hydrolase domain | tRNA-Arg | DNA-binding transcriptional repressor | bicyclomycin/multidrug efflux system | cyanate aminohydrolase | DLP12 prophage; multidrug resistance protein | tRNA-Leu | CP4-6 prophage; S-methylmethionine:homocysteine methyltransferase | regulatory protein for HokC, overlaps CDS of hokC | phenylalanine transporter | gamma-glutamylphosphate reductase | tRNA-Pro | tRNA(Ile)-lysidine synthetase | CP4-6 prophage; predicted lyase/synthase | CP4-6 prophage; predicted DNA-binding transcriptional regulator | predicted hydratase | predicted hydrolase, inner membrane | predicted NAD(P)H-binding oxidoreductase with NAD(P)-binding Rossmann-fold domain | predicted inner membrane protein | predicted enzyme | predicted ATP-dependet helicase | predicted NAD(P)-binding oxidoreductase with NAD(P)-binding Rossmann-fold domain | predicted peptide transporter subunit: membrane component of ABC superfamily |
| 116 | 74 | 338 | 0.963 | 0.547 |  |  |  |  |  | b1927 b0123 b4314 b4317 b4318 b4319 b4315 b0912 b2127 b4430 b4441 b0480 b0581 b0773 b0767 b0925 b2379 b2449 b1450 b2475 b0049 b2079 b1661 b0882 b0881 b2417 b0880 b4408 b3234 b2364 b2441 b4316 b4320 b2804 b0811 b1285 b1064 b1619 b0047 b0046 b0870 b2687 b2074 b0957 b2599 b2598 b0708 b1353 b0494 b0412 b0454 b0495 b0713 b0843 b0869 b0966 b4492 b1895 b2008 b2015 b2190 b2304 b2347 b2448 b2627 b2671 b2749 b2989 b0987 b4525 b1810 b1957 b1998 b3102 | amyA cueO fimA fimD fimF fimG fimI ihfB mlrA ushA ybdK ybhB pgl ycbB yfdZ yffR yncC ypfJ apaH baeR cfa clpA clpS crr cspD degQ dsdC eutB fimC fimH fucU glnH gmr grxB hdhA kefC kefF ltaE luxS mdtA ompA pheA pheL phr sieB tesA yajI ybaZ ybbA ybgL ybjH ybjT hspQ ydbA uspC yeeA yeeY yejO yfcH yfdC yffQ yfjK ygaC ygbE yghU gfcA ymjC yoaC yodC yqjG | cytoplasmic alpha-amylase | multicopper oxidase (laccase) | major type 1 subunit fimbrin (pilin) | outer membrane usher protein, type 1 fimbrial synthesis | minor component of type 1 fimbriae | fimbrial protein involved in type 1 pilus biosynthesis | integration host factor (IHF), DNA-binding protein, beta subunit | DNA-binding transcriptional regulator | bifunctional UDP-sugar hydrolase/5'-nucleotidase | gamma-glutamyl:cysteine ligase | predicted kinase inhibitor | 6-phosphogluconolactonase | predicted carboxypeptidase | prediected aminotransferase, PLP-dependent | CPZ-55 prophage; predicted protein | predicted DNA-binding transcriptional regulator | conserved protein | diadenosine tetraphosphatase | DNA-binding response regulator in two-component regulatory system with BaeS | cyclopropane fatty acyl phospholipid synthase (unsaturated-phospholipid methyltransferase) | ATPase and specificity subunit of ClpA-ClpP ATP-dependent serine protease, chaperone activity | regulatory protein for ClpA substrate specificity | glucose-specific enzyme IIA component of PTS | cold shock protein homolog | serine endoprotease, periplasmic | DNA-binding transcriptional dual regulator | ethanolamine ammonia-lyase, large subunit, heavy chain | chaperone, periplasmic | L-fucose mutarotase | glutamine transporter subunit | modulator of Rnase II stability | glutaredoxin 2 (Grx2) | 7-alpha-hydroxysteroid dehydrogenase, NAD-dependent | potassium:proton antiporter | flavoprotein subunit for the KefC potassium efflux system | L-allo-threonine aldolase, PLP-dependent | S-ribosylhomocysteinase | multidrug efflux system, subunit A | outer membrane protein A (3a;II*;G;d) | fused chorismate mutase P/prephenate dehydratase | pheA gene leader peptide | deoxyribodipyrimidine photolyase, FAD-binding | Rac prophage; phage superinfection exclusion protein | multifunctional acyl-CoA thioesterase I and protease I and lysophospholipase L1 | predicted lipoprotein | predicted methyltransferase | predicted transporter subunit: ATP-binding component of ABC superfamily | predicted lactam utilization protein | predicted protein | conserved protein with NAD(P)-binding Rossmann-fold domain | DNA-binding protein, hemimethylated | universal stress protein | conserved inner membrane protein | predicted inner membrane protein | CP4-57 prophage; conserved protein | predicted S-transferase |
| 34 | 81 | 337 | 1.075 | 0.600 |  |  | 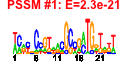 | 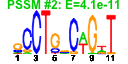 |  | b4150 b4401 b3237 b0313 b2810 b3472 b4124 b4125 b0145 b0460 b4271 b3350 b3351 b4386 b0448 b0192 b3360 b3775 b3748 b3794 b4179 b2572 b2571 b2570 b4456 b0097 b3349 b2617 b4148 b3273 b4501 b3761 b3793 b0119 b0191 b4406 b0190 b2668 b2667 b2811 b2833 b3021 b3107 b3353 b4551 b3467 b3468 b3795 b3928 b4178 b4255 b4385 b2666 b4209 b2910 b3530 b0311 b0312 b0314 b4484 b3912 b3915 b4172 b0096 b4171 b2159 b0019 b3790 b0189 b2573 b0461 b0447 b2160 b2795 b2865 b3884 b4011 b4061 b4402 b4505 b3101 | ampC arcA argR betI csdA dcrB dcuR dcuS dksA hha intD kefB kefG lplA mdlA nlpE pabA ppiC rbsD rffM rnr rseA rseB rseC secM slyD smpA sugE thrV torI trpT wzyE yacL yaeJ yaeP yaeQ ygaP ygaV csdE ygdR ygiT yhaL yheT yheV yhhM yhhN yifK yiiU nsrR rraB yjjJ yqaE ytfE zapA bcsC betA betB betT cpxP cpxR fieF hfq lpxC miaA nfo nhaA rffC rof rpoE ybaJ ybaO yeiI ygdH ygeR yihW yjaA yjcC yjjY ykgN yqjF | beta-lactamase/D-alanine carboxypeptidase | DNA-binding response regulator in two-component regulatory system with ArcB or CpxA | DNA-binding transcriptional dual regulator, L-arginine-binding | DNA-binding transcriptional repressor | cysteine sulfinate desulfinase | periplasmic protein | DNA-binding response regulator in two-component regulatory system with DcuS | sensory histidine kinase in two-component regulatory system with DcuR, regulator of anaerobic fumarate respiration | DNA-binding transcriptional regulator of rRNA transcription, DnaK suppressor protein | modulator of gene expression, with H-NS | DLP12 prophage; predicted integrase | potassium:proton antiporter | component of potassium effux complex with KefB | lipoate-protein ligase A | fused predicted multidrug transporter subunits of ABC superfamily: ATP-binding components | lipoprotein involved with copper homeostasis and adhesion | aminodeoxychorismate synthase, subunit II | peptidyl-prolyl cis-trans isomerase C (rotamase C) | predicted cytoplasmic sugar-binding protein | UDP-N-acetyl-D-mannosaminuronic acid transferase | exoribonuclease R, RNase R | anti-sigma factor | RseC protein involved in reduction of the SoxR iron-sulfur cluster | regulator of secA translation | FKBP-type peptidyl prolyl cis-trans isomerase (rotamase) | small membrane lipoprotein | multidrug efflux system protein | tRNA-Thr | response regulator inhibitor for tor operon | tRNA-Trp | predicted Wzy protein involved in ECA polysaccharide chain elongation | conserved protein | predicted inner membrane protein with hydrolase activity | predicted DNA-binding transcriptional regulator | predicted Fe-S metabolism protein | predicted protein | predicted hydrolase | conserved inner membrane protein | predicted transporter | predicted membrane protein | predicted regulator of cell morphogenesis and cell wall metabolism | protein that localizes to the cytokinetic ring | cellulose synthase subunit | choline dehydrogenase, a flavoprotein | betaine aldehyde dehydrogenase, NAD-dependent | choline transporter of high affinity | periplasmic protein combats stress | DNA-binding response regulator in two-component regulatory system with CpxA | zinc transporter | HF-I, host factor for RNA phage Q beta replication | UDP-3-O-acyl N-acetylglucosamine deacetylase | delta(2)-isopentenylpyrophosphate tRNA-adenosine transferase | endonuclease IV with intrinsic 3'-5' exonuclease activity | sodium-proton antiporter | TDP-fucosamine acetyltransferase | modulator of Rho-dependent transcription termination | RNA polymerase, sigma 24 (sigma E) factor | predicted kinase | tetratricopeptide repeat transcriptional regulator | predicted signal transduction protein (EAL domain containing protein) | predicted IS protein | predicted quinol oxidase subunit |
| 57 | 70 | 330 | 1.128 | 0.567 |  |  | 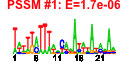 | 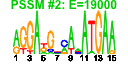 |  | b4408 b2367 b2492 b0124 b2373 b2519 b0556 b0316 b0562 b0599 b0601 b0602 b1309 b1963 b1972 b2375 b2443 b2446 b2520 b2754 b2755 b2756 b2757 b2758 b2759 b2760 b3268 b0303 b4520 b2376 b3014 b0516 b2370 b2374 b2883 b1182 b0076 b3828 b4427 b0383 b4433 b2621 b2867 b2868 b0277 b0500 b0821 b2083 b2305 b2371 b2420 b2444 b2445 b2561 b2629 b2633 b2668 b2667 b2833 b0300 b0304 b0305 b0288 b0392 b0499 b1167 b4524 b1551 b2550 b2639 | emrY focB gcd oxc pbpC rzpD yahB ybcY ybdH ybdM ybdN ycjM yedR yedZ yfdX yffL yffO yfhM ygbF ygbT ygcH ygcI ygcJ ygcK ygcL yhdW ykgI ymgF ypdI yqhH allC evgS frc guaD hlyE leuO metR phoA xdhB xdhC yagK ybbD ybiU yegZ yfcI yfdE yfeS yffM yffN yfhH yfjM yfjQ ygaP ygaV ygdR ykgA ykgC ykgD ykgJ ykiA ylbH ymgC ycjV ynfN yphH | predicted multidrug efflux system | predicted formate transporter | glucose dehydrogenase | predicted oxalyl-CoA decarboxylase | fused transglycosylase/transpeptidase | DLP12 prophage; predicted murein endopeptidase | predicted DNA-bindng transcriptional regulator | predicted oxidoreductase | conserved protein | predicted glucosyltransferase | predicted inner membrane protein | conserved inner membrane protein | predicted protein | CPZ-55 prophage; predicted protein | predicted lipoprotein involved in colanic acid biosynthesis | predicted outer membrane lipoprotein | allantoate amidohydrolase | hybrid sensory histidine kinase in two-component regulatory system with EvgA | formyl-CoA transferase, NAD(P)-binding | guanine deaminase | hemolysin E | DNA-binding transcriptional activator | DNA-binding transcriptional activator, homocysteine-binding | bacterial alkaline phosphatase | xanthine dehydrogenase, FAD-binding subunit | xanthine dehydrogenase, Fe-S binding subunit | CP4-6 prophage; conserved protein | predicted CoA-transferase, NAD(P)-binding | predicted DNA-binding transcriptional regulator | CP4-57 prophage; predicted protein | predicted inner membrane protein with hydrolase activity | predicted oxidoreductase with FAD/NAD(P)-binding domain and dimerization domain | predicted ferredoxin | Qin prophage; predicted protein |
| 81 | 75 | 339 | 1.201 | 0.578 |  |  | 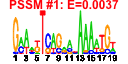 | 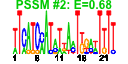 |  | b0062 b0390 b3536 b3537 b3538 b4225 b0155 b3634 b0103 b4397 b3182 b3058 b3924 b4140 b3672 b2843 b2701 b0020 b3405 b3977 b0056 b0122 b0478 b1060 b2777 b2928 b2929 b3219 b3535 b3713 b3872 b3910 b4065 b4403 b3095 b4220 b4221 b0388 b2783 b3911 b4398 b2810 b4124 b0145 b2808 b0460 b4271 b3633 b2781 b3171 b3742 b4219 b1109 b3397 b2082 b4041 b2972 b3842 b4179 b4456 b4501 b4241 b4039 b2604 b2811 b2971 b4466 b3574 b3795 b3928 b4178 b4255 b4262 b4222 b2910 | araA aroM bcsE bcsF bcsG chpB clcA coaD coaE creA dacB folB fpr fxsA ivbL kduI mltB nhaR ompR tyrU yacC ybaL bssS ygcF yggC yggD yhcF yhjR yieF yihL yiiM yjcE yjtD yqjA ytfM ytfN aroL chpR cpxA creB csdA dcuR dksA gcvA hha intD waaA mazG metY mioC msrA ndh nudE ogrK plsB pppA rfaH rnr torI treR ubiC yfiN csdE yghG yghJ yiaJ yifK yiiU nsrR rraB yjgQ ytfP zapA | L-arabinose isomerase | conserved protein | predicted protein | predicted inner membrane protein | toxin of the ChpB-ChpS toxin-antitoxin system | chloride channel, voltage-gated | pantetheine-phosphate adenylyltransferase | dephospho-CoA kinase | D-alanyl-D-alanine carboxypeptidase | bifunctional dihydroneopterin aldolase/dihydroneopterin triphosphate 2'-epimerase | ferredoxin-NADP reductase | inner membrane protein | ilvB operon leader peptide | predicted 5-keto 4-deoxyuronate isomerase | membrane-bound lytic murein transglycosylase B | DNA-binding transcriptional activator | DNA-binding response regulator in two-component regulatory system with EnvZ | tRNA-Tyr | predicted transporter with NAD(P)-binding Rossmann-fold domain | conserved protein with nucleoside triphosphate hydrolase domain | predicted DNA-binding transcriptional regulator | predicted transcriptional regulator | chromate reductase, Class I, flavoprotein | predicted cation/proton antiporter | predicted rRNA methyltransferase | conserved inner membrane protein | predicted outer membrane protein and surface antigen | shikimate kinase II | antitoxin of the ChpA-ChpR toxin-antitoxin system | sensory histidine kinase in two-component regulatory system with CpxR | DNA-binding response regulator in two-component regulatory system with CreC | cysteine sulfinate desulfinase | DNA-binding response regulator in two-component regulatory system with DcuS | DNA-binding transcriptional regulator of rRNA transcription, DnaK suppressor protein | DNA-binding transcriptional dual regulator | modulator of gene expression, with H-NS | DLP12 prophage; predicted integrase | 3-deoxy-D-manno-octulosonic-acid transferase (KDO transferase) | nucleoside triphosphate pyrophosphohydrolase | tRNA-Met | FMN-binding protein MioC | methionine sulfoxide reductase A | respiratory NADH dehydrogenase 2/cupric reductase | ADP-ribose diphosphatase | DNA-binding transcriptional regulator prophage P2 remnant | glycerol-3-phosphate O-acyltransferase | bifunctional prepilin leader peptidase/ methylase | DNA-binding transcriptional antiterminator | exoribonuclease R, RNase R | response regulator inhibitor for tor operon | DNA-binding transcriptional repressor | chorismate pyruvate lyase | predicted diguanylate cyclase | predicted Fe-S metabolism protein | predicted inner membrane lipoprotein | predicted DNA-binding transcriptional repressor | predicted transporter | protein that localizes to the cytokinetic ring |
| 98 | 79 | 330 | 1.216 | 0.545 |  |  | 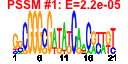 | 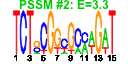 |  | b3243 b4498 b4120 b3599 b3025 b4449 b0217 b0322 b2645 b2857 b2859 b2921 b3173 b3369 b3488 b3584 b3585 b3594 b4562 b0310 b0294 b1151 b2847 b2848 b3241 b3242 b4115 b3133 b4400 b4313 b3989 b4285 b2842 b0448 b4094 b3026 b4063 b4062 b4148 b0008 b0007 b0193 b0164 b0539 b0629 b0705 b4514 b2956 b2968 b2969 b2970 b2981 b2998 b3060 b3023 b3090 b3154 b3155 b3219 b3817 b3878 b4020 b4029 b4559 b4231 b4183 b4249 b4299 b4309 b4336 b4338 b0235 b0303 b1152 b2657 b2846 b3014 b3446 b4211 | aaeR gatR melB mtlA qseB yafT yfjZ ygeO ygfI yhbX yhfL yhiJ yiaT yiaU yibA ykgH matA ymfO yqeI yqeJ aaeA aaeX adiC agaV creD fimE yjaZ kduD mdlA phnN qseC soxR soxS sugE talB yaaJ yaeF yaeI ybcC ybeF ybfL ybfQ yggM yghD yghE yghF yghO yghW ttdR ygiV ygjV yhbP yhbQ yhcF yigF yihQ yjbB yjbH yjdO yjfF yjfK yjgI yjhI yjhS yjiN ykfJ ykgI ymfP yqeH yqhH yrhB ytfG | predicted DNA-binding transcriptional regulator, efflux system | melibiose:sodium symporter | fused mannitol-specific PTS enzymes: IIA components/IIB components/IIC components | DNA-binding response regulator in two-component regulatory system with QseC | predicted aminopeptidase | CP4-57 prophage; antitoxin of the YpjF-YfjZ toxin-antitoxin system | predicted DNA-binding transcriptional regulator | predicted hydrolase, inner membrane | conserved secreted peptide | predicted protein | lyase containing HEAT-repeat | predicted inner membrane protein | predicted regulator | predicted transcriptional regulator | p-hydroxybenzoic acid efflux system component | membrane protein of efflux system | arginine:agmatin | N-acetylgalactosamine-specific enzyme IIB component of PTS | inner membrane protein | tyrosine recombinase/inversion of on/off regulator of fimA | heat shock protein | 2-deoxy-D-gluconate 3-dehydrogenase | fused predicted multidrug transporter subunits of ABC superfamily: ATP-binding components | ribose 1,5-bisphosphokinase | sensory histidine kinase in two-component regulatory system with QseB | DNA-binding transcriptional dual regulator, Fe-S center for redox-sensing | DNA-binding transcriptional dual regulator | multidrug efflux system protein | transaldolase B | predicted transporter | predicted lipoprotein | predicted phosphatase | predicted transposase | conserved protein | predicted secretion pathway M-type protein, membrane anchored | conserved inner membrane protein | predicted endonuclease | alpha-glucosidase | predicted porin | predicted sugar transporter subunit: membrane component of ABC superfamily | predicted oxidoreductase with NAD(P)-binding Rossmann-fold domain | KpLE2 phage-like element; predicted DNA-binding transcriptional regulator | conserved protein with bipartite regulator domain | predicted outer membrane lipoprotein | NAD(P)H:quinone oxidoreductase |
| 137 | 68 | 340 | 1.229 | 0.569 |  |  |  |  |  | b0842 b2140 b0503 b0885 b2686 b2803 b1550 b0849 b2538 b4415 b0650 b1379 b2349 b2411 b1139 b4570 b1532 b0852 b4437 b2353 b2324 b2582 b2028 b0127 b0128 b0270 b0271 b0528 b0691 b0846 b0847 b0959 b1021 b1022 b1023 b1024 b1025 b1055 b1366 b1422 b1446 b1497 b1498 b1504 b1572 b1649 b1785 b1797 b2085 b2172 b2181 b2250 b2343 b2395 b2409 b2581 b2642 b2776 b2930 b4028 b0457 b1028 b1122 b1171 b4542 b4546 b2641 b0752 | cmr dusC ybbB aat emrB fucK gnsB grxA hcaE hokE hscC hslJ intS ligA lit lomR marB rimK tfaS mnmC trxC ugd yadG yadH yagG yagH ybcJ ybfG ybjK ybjL sxy pgaD pgaC pgaB pgaA ycdT yceA ydaY ydcI ydcY ydeM ydeN ydeS ydfB ydhM yeaI yeaR yegR yeiQ yejG yfaZ yfcZ yfeA yfeR yfiF yfjW ygcE yggF yjbG ylaB ymdE ymfA ymgD yohO ypeB yfjV zitB | multidrug efflux system protein | tRNA-dihydrouridine synthase C | tRNA 2-selenouridine synthase, selenophosphate-dependent | leucyl/phenylalanyl-tRNA-protein transferase | L-fuculokinase | Qin prophage; predicted protein | glutaredoxin 1, redox coenzyme for ribonucleotide reductase (RNR1a) | 3-phenylpropionate dioxygenase, large (alpha) subunit | toxic polypeptide, small | Hsp70 family chaperone Hsc62, binds to RpoD and inhibits transcription | heat-inducible protein | CPS-53 (KpLE1) prophage; predicted prophage CPS-53 integrase | DNA ligase, NAD(+)-dependent | e14 prophage; cell death peptidase, inhibitor of T4 late gene expression | predicted protein | ribosomal protein S6 modification protein | fused 5-methylaminomethyl-2-thiouridine-forming enzyme methyltransferase/FAD-dependent demodification enzyme | thioredoxin 2 | UDP-glucose 6-dehydrogenase | predicted transporter subunit: ATP-binding component of ABC superfamily | predicted transporter subunit: membrane component of ABC superfamily | CP4-6 prophage; predicted sugar transporter | CP4-6 prophage; predicted xylosidase/arabinosidase | predicted RNA-binding protein | predicted DNA-binding transcriptional regulator | predicted transporter | conserved protein | predicted inner membrane protein | predicted glycosyl transferase | predicted enzyme associated with biofilm formation | predicted outer membrane protein | predicted diguanylate cyclase | predicted fimbrial-like adhesin protein | predicted dehydrogenase, NAD-dependent | predicted outer membrane porin protein | predicted methyltransferase | CP4-57 prophage; predicted inner membrane protein | predicted kinase | predicted hexoseP phosphatase | conserved inner membrane protein | zinc efflux system |
| 13 | 69 | 328 | 1.276 | 0.582 |  |  |  |  |  | b3135 b3136 b3134 b3744 b3533 b3531 b3717 b0162 b4399 b4400 b3370 b3371 b4474 b3682 b3437 b2536 b3391 b2237 b3137 b4453 b3647 b0449 b1599 b3161 b3935 b3642 b3026 b3903 b3668 b4195 b4197 b3564 b0130 b0315 b0456 b1821 b2238 b2534 b2586 b2640 b2806 b2969 b3063 b3471 b3589 b3638 b3675 b3716 b3719 b3827 b3876 b3877 b3890 b4029 b4121 b4286 b4327 b4332 b3395 b4556 b4210 b3532 b3415 b4476 b3940 b3666 b0024 b3434 b3469 | agaA agaS agaW asnA bcsA bcsZ cbrC cdaR creC creD frlA frlB frlC gntK hcaT hofQ inaA kbaY ldrD ligB mdlB mdtI mtr priA pyrE qseC rhaA uhpB ulaC ulaE xylB yadE yahA ybaA yebN yfaH yfhR yfiM ygdE yghE ttdT yhhQ yiaY yicR yidG cbrB yieL yigM yihO yihP yiiF yjbH yjdF yjhV yjiE yjiJ hofM glvG ytfF bcsB gntT gntU metL uhpT yaaY yhgN zntA | tagatose-6-phosphate ketose/aldose isomerase | asparagine synthetase A | cellulose synthase, catalytic subunit | endo-1,4-D-glucanase | conserved protein | DNA-binding transcriptional activator | sensory histidine kinase in two-component regulatory system with CreB or PhoB, regulator of the CreBC regulon | inner membrane protein | predicted fructoselysine transporter | fructoselysine-6-P-deglycase | predicted isomerase | gluconate kinase 2 | predicted 3-phenylpropionic transporter | predicted fimbrial transporter | tagatose 6-phosphate aldolase 1, kbaY subunit | toxic polypeptide, small | DNA ligase, NAD(+)-dependent | fused predicted multidrug transporter subunits of ABC superfamily: ATP-binding components | multidrug efflux system transporter | tryptophan transporter of high affinity | Primosome factor n' (replication factor Y) | orotate phosphoribosyltransferase | sensory histidine kinase in two-component regulatory system with QseB | L-rhamnose isomerase | sensory histidine kinase in two-component regulatory sytem with UhpA | L-ascorbate-specific enzyme IIA component of PTS | L-xylulose 5-phosphate 3-epimerase | xylulokinase | predicted polysaccharide deacetylase lipoprotein | predicted DNA-binding transcriptional regulator | conserved inner membrane protein | predicted peptidase | predicted protein | predicted methyltransferase | predicted tartrate:succinate antiporter | predicted Fe-containing alcohol dehydrogenase | protein associated with replication fork, possible DNA repair protein | predicted inner membrane protein | predicted xylanase | predicted transporter | predicted porin | predicted pilus assembly protein | regulator of cellulose synthase, cyclic di-GMP binding | gluconate transporter, high-affinity GNT I system | gluconate transporter, low affinity GNT 1 system | fused aspartokinase II/homoserine dehydrogenase II | hexose phosphate transporter | predicted antibiotic transporter | zinc, cobalt and lead efflux system |
| 124 | 90 | 332 | 1.362 | 0.587 |  |  |  |  |  | b2708 b2733 b1660 b3292 b0063 b0061 b2841 b0396 b2923 b0390 b3338 b4225 b3556 b1557 b1558 b0990 b0989 b1552 b0231 b3645 b4362 b1160 b2943 b2706 b1140 b2843 b4043 b1600 b2701 b2831 b0099 b0020 b0113 b0060 b3025 b3819 b2616 b2698 b1861 b1860 b4445 b2704 b2559 b3115 b1184 b1183 b4058 b0779 b0233 b0234 b0217 b0848 b1060 b1457 b1728 b1846 b1848 b1968 b2680 b2777 b2832 b2840 b2857 b2859 b2863 b2912 b3009 b3036 b4550 b3293 b3547 b3660 b4060 b4128 b4575 b4342 b1141 b1142 b1144 b1147 b1148 b1151 b1150 b1146 b4520 b1458 b1459 b2847 b2848 b4218 | gutQ mutS ydhC zntR araB araD araE araJ argO aroM chiA chpB cspA cspB cspF cspG cspH cspI dinB dinD dnaT elbA galP gutM intE kduI lexA mdtJ mltB mutH mutT nhaR pdhR polB qseB rarD recN recX ruvA ruvB srlB tadA tdcD umuC umuD uvrA uvrB yafO yafP yafT ybjM bssS ydcD ydjM yebE yebG yedV ygcF ygdQ ygeA ygeO ygeQ ygfA yghB yhdL yhdN yhjX yicL yjcB yjdK yjgX yjiT xisE ymfJ ymfL ymfM ymfO ymfR ymfT ymgF yncI yqeI yqeJ ytfL | predicted phosphosugar-binding protein | methyl-directed mismatch repair protein | predicted transporter | DNA-binding transcriptional activator in response to Zn(II) | L-ribulokinase | L-ribulose-5-phosphate 4-epimerase | arabinose transporter | arginine transporter | conserved protein | periplasmic endochitinase | toxin of the ChpB-ChpS toxin-antitoxin system | major cold shock protein | Qin prophage; cold shock protein | DNA-binding transcriptional regulator | stress protein, member of the CspA-family | DNA polymerase IV | DNA-damage-inducible protein | DNA biosynthesis protein (primosomal protein I) | predicted protein | D-galactose transporter | DNA-binding transcriptional activator of glucitol operon | e14 prophage; predicted integrase | predicted 5-keto 4-deoxyuronate isomerase | DNA-binding transcriptional repressor of SOS regulon | multidrug efflux system transporter | membrane-bound lytic murein transglycosylase B | nucleoside triphosphate pyrophosphohydrolase, marked preference for dGTP | DNA-binding transcriptional activator | DNA-binding transcriptional dual regulator | DNA polymerase II | DNA-binding response regulator in two-component regulatory system with QseC | predicted chloramphenical resistance permease | recombination and repair protein | regulatory protein for RecA | component of RuvABC resolvasome, regulatory subunit | ATP-dependent DNA helicase, component of RuvABC resolvasome | glucitol/sorbitol-specific enzyme IIA component of PTS | tRNA-specific adenosine deaminase | propionate kinase/acetate kinase C, anaerobic | DNA polymerase V, subunit C | DNA polymerase V, subunit D | ATPase and DNA damage recognition protein of nucleotide excision repair excinuclease UvrABC | excinulease of nucleotide excision repair, DNA damage recognition component | predicted toxin of the YafO-YafN toxin-antitoxin system | predicted acyltransferase with acyl-CoA N-acyltransferase domain | predicted aminopeptidase | predicted inner membrane protein | predicted inner membrane protein regulated by LexA | conserved protein regulated by LexA | predicted sensory kinase in two-component regulatory system with YedW | predicted racemase | predicted ligase | conserved inner membrane protein | e14 prophage; predicted excisionase | e14 prophage; predicted protein | e14 prophage; predicted DNA-binding transcriptional regulator | predicted transcriptional regulator |
| 1 | 84 | 337 | 1.508 | 0.653 |  |  |  |  |  | b2435 b3057 b0311 b0312 b0314 b3337 b3966 b0158 b0381 b0161 b0160 b4136 b3006 b3005 b4289 b4288 b4293 b4292 b3408 b3409 b0150 b0153 b0151 b0152 b3515 b2903 b3053 b3414 b0059 b2527 b3055 b2528 b2531 b2530 b2529 b4434 b2587 b0186 b1380 b1600 b3940 b4499 b0099 b2159 b0814 b0932 b4155 b4111 b4451 b3908 b4448 b3553 b1907 b3669 b0006 b0144 b0187 b0468 b0631 b1112 b1378 b2160 b2187 b2251 b2561 b2562 b4461 b3009 b3037 b3038 b3029 b3064 b3410 b3434 b4061 b4543 b2380 b2381 b2434 b4547 b2790 b2845 b2899 b3427 | amiA bacA betA betB betT bfd btuB btuF ddlA degP dgt dipZ exbB exbD fecC fecD fecI fecR feoA feoB fhuA fhuB fhuC fhuD gadW gcvP glnE gntY hepA hscB htrG iscA iscR iscS iscU kgtP ldcC ldhA mdtJ metL yehH mutT nfo ompX pepN poxA proP sodA ghrB tyrP uhpA yaaA yadB yaeR ybaN ybeD bhsA ydbK yeiI yejL nudI yfhH yfhL yfjD yghB ygiB ygiC ygiN ygjD feoC yhgN yjcC ypaA ypdA ypdB ypeA ypfN yqcA yqeG yqfA yzgL | N-acetylmuramoyl-l-alanine amidase I | undecaprenyl pyrophosphate phosphatase | choline dehydrogenase, a flavoprotein | betaine aldehyde dehydrogenase, NAD-dependent | choline transporter of high affinity | bacterioferritin-associated ferredoxin | vitamin B12/cobalamin outer membrane transporter | vitamin B12 transporter subunit: periplasmic-binding component of ABC superfamily | D-alanine-D-alanine ligase A | serine endoprotease (protease Do), membrane-associated | deoxyguanosine triphosphate triphosphohydrolase | fused thiol:disulfide interchange protein: activator of DsbC/conserved protein | membrane spanning protein in TonB-ExbB-ExbD complex | KpLE2 phage-like element; iron-dicitrate transporter subunit | KpLE2 phage-like element; RNA polymerase, sigma 19 factor | KpLE2 phage-like element; transmembrane signal transducer for ferric citrate transport | ferrous iron transporter, protein A | fused ferrous iron transporter, protein B: GTP-binding protein/membrane protein | ferrichrome outer membrane transporter | fused iron-hydroxamate transporter subunits of ABC superfamily: membrane components | iron-hydroxamate transporter subunit | DNA-binding transcriptional activator | glycine decarboxylase, PLP-dependent, subunit (protein P) of glycine cleavage complex | fused deadenylyltransferase/adenylyltransferase for glutamine synthetase | predicted gluconate transport associated protein | RNA polymerase-associated helicase protein (ATPase and RNA polymerase recycling factor) | DnaJ-like molecular chaperone specific for IscU | predicted signal transduction protein (SH3 domain) | FeS cluster assembly protein | DNA-binding transcriptional repressor | cysteine desulfurase (tRNA sulfurtransferase), PLP-dependent | scaffold protein | alpha-ketoglutarate transporter | lysine decarboxylase 2, constitutive | fermentative D-lactate dehydrogenase, NAD-dependent | multidrug efflux system transporter | fused aspartokinase II/homoserine dehydrogenase II | nucleoside triphosphate pyrophosphohydrolase, marked preference for dGTP | endonuclease IV with intrinsic 3'-5' exonuclease activity | outer membrane protein | aminopeptidase N | predicted lysyl-tRNA synthetase | proline/glycine betaine transporter | superoxide dismutase, Mn | 2-keto-D-gluconate reductase (glyoxalate reductase) (2-ketoaldonate reductase) | tyrosine transporter | DNA-binding response regulator in two-component regulatory system wtih UhpB | conserved protein | glutamyl-Q tRNA(Asp) synthetase | predicted lyase | conserved inner membrane protein | predicted protein | fused predicted pyruvate-flavodoxin oxidoreductase: conserved protein/conserved protein/FeS binding protein | predicted kinase | predicted NUDIX hydrolase | predicted DNA-binding transcriptional regulator | predicted 4Fe-4S cluster-containing protein | predicted inner membrane protein | conserved outer membrane protein | predicted enzyme | quinol monooxygenase | predicted peptidase | predicted antibiotic transporter | predicted signal transduction protein (EAL domain containing protein) | predicted sensory kinase in two-component system with YpdB | predicted response regulator in two-component system withYpdA | predicted acyltransferase with acyl-CoA N-acyltransferase domain | predicted flavoprotein | predicted transporter | predicted oxidoreductase, inner membrane subunit |
| 8 | 74 | 335 | 1.564 | 0.600 |  |  |  |  |  | b2470 b3072 b3503 b4112 b3532 b4213 b4208 b4214 b3227 b3673 b4114 b2980 b3945 b4321 b3415 b4476 b0106 b0012 b4265 b2842 b4331 b2838 b3513 b3514 b1053 b4019 b3828 b0260 b2574 b4073 b3824 b4090 b3855 b4302 b4301 b2965 b3089 b3452 b3451 b3449 b3666 b0010 b0007 b0005 b0379 b1797 b2389 b2835 b3040 b3160 b3263 b3507 b3524 b3548 b3654 b3943 b4030 b4020 b4060 b4064 b4078 b4190 b4334 b4363 b4364 b0250 b0288 b0295 b0296 b2917 b2966 b3151 b3469 b3039 | acrD aer arsC basS bcsB cpdB cycA cysQ dcuD emrD eptA glcC gldA gntP gntT gntU hofC idnT kduD kptA lysA mdtE mdtF mdtG metH metR mmuP nadB nrfD rhtB rpiB rrfA sgcA sgcE speC sstT ugpA ugpE ugpQ uhpT yaaH yaaJ yaaX yaiY yeaR yfeO lplT zupT yhbW yhdU dctR yhjG yhjY yicE yijE psiE yjbB yjcB yjcD yjcO yjfP yjiL yjjB yjjP ykfB ykgJ ykgL ykgM scpA yqgA yraQ zntA ygiD | aminoglycoside/multidrug efflux system | fused signal transducer for aerotaxis sensory component/methyl accepting chemotaxis component | arsenate reductase | sensory histidine kinase in two-component regulatory system with BasR | regulator of cellulose synthase, cyclic di-GMP binding | 2':3'-cyclic-nucleotide 2'-phosphodiesterase | D-alanine/D-serine/glycine transporter | PAPS (adenosine 3'-phosphate 5'-phosphosulfate) 3'(2'),5'-bisphosphate nucleotidase | predicted transporter | multidrug efflux system protein | predicted metal dependent hydrolase | DNA-binding transcriptional dual regulator, glycolate-binding | glycerol dehydrogenase, NAD | fructuronate transporter | gluconate transporter, high-affinity GNT I system | gluconate transporter, low affinity GNT 1 system | assembly protein in type IV pilin biogenesis, transmembrane protein | L-idonate and D-gluconate transporter | 2-deoxy-D-gluconate 3-dehydrogenase | 2'-phosphotransferase | diaminopimelate decarboxylase, PLP-binding | multidrug resistance efflux transporter | multidrug transporter, RpoS-dependent | predicted drug efflux system | homocysteine-N5-methyltetrahydrofolate transmethylase, B12-dependent | DNA-binding transcriptional activator, homocysteine-binding | CP4-6 prophage; predicted S-methylmethionine transporter | quinolinate synthase, L-aspartate oxidase (B protein) subunit | formate-dependent nitrite reductase, membrane subunit | neutral amino-acid efflux system | ribose 5-phosphate isomerase B/allose 6-phosphate isomerase | 5S ribosomal RNA of rrnA operon | KpLE2 phage-like element; predicted phosphotransferase enzyme IIA component | KpLE2 phage-like element; predicted epimerase | ornithine decarboxylase, constitutive | sodium:serine/threonine symporter | glycerol-3-phosphate transporter subunit | glycerophosphodiester phosphodiesterase, cytosolic | hexose phosphate transporter | conserved inner membrane protein associated with acetate transport | predicted protein | predicted inner membrane protein | conserved protein | predicted ion channel protein | zinc transporter | predicted enzyme | predicted membrane protein | predicted DNA-binding ranscriptional regulator | predicted outer membrane biogenesis protein | predicted permease | predicted phosphate starvation inducible protein | predicted hydrolase | predicted ATPase, activator of (R)-hydroxyglutaryl-CoA dehydratase | conserved inner membrane protein | CP4-6 prophage; predicted protein | predicted ferredoxin | rpmJ (L36) paralog | methylmalonyl-CoA mutase | zinc, cobalt and lead efflux system | predicted dioxygenase |
| 52 | 71 | 326 | 1.569 | 0.606 |  |  |  |  |  | b0536 b2348 b3796 b3022 b2417 b1910 b4362 b2864 b3438 b2496 b3797 b2652 b4294 b4369 b4270 b1909 b3171 b4104 b4094 b3545 b2189 b3799 b2416 b4431 b4433 b3658 b0971 b2695 b4429 b2621 b0244 b3450 b1665 b0156 b0327 b0966 b1571 b1667 b2575 b3439 b0987 b0991 b1167 b4542 b3744 b0039 b4224 b0161 b0455 b0255 b4095 b4092 b3123 b3855 b4417 b4444 b4446 b1975 b0070 b4418 b3668 b0065 b0276 b0802 b1378 b2586 b3471 b3888 b4026 b4547 b4223 | argU argW argX mqsR crr cysU dnaT glyU gntR hda hisR ileY insA leuP leuX leuZ metY phnE phnN proK proL proM ptsI serT serV thrW ugpC valV yadR yahM hspQ ydfA ydhR yfiC yhhW gfcA ymcE ymgC yohO asnA caiA chpS degP phnM phnP rrfA serU setA uhpB yabI yagJ ybiJ ydbK yfiM yhhQ yiiD yjbE ypfN | tRNA-Arg | predicted cyanide hydratase | glucose-specific enzyme IIA component of PTS | sulfate/thiosulfate transporter subunit | DNA biosynthesis protein (primosomal protein I) | tRNA-Gly | DNA-binding transcriptional repressor | ATPase regulatory factor involved in DnaA inactivation | tRNA-His | tRNA-Ile | KpLE2 phage-like element; IS1 repressor protein InsA | tRNA-Leu | tRNA-Met | ribose 1,5-bisphosphokinase | tRNA-Pro | PEP-protein phosphotransferase of PTS system (enzyme I) | tRNA-Ser | tRNA-Thr | glycerol-3-phosphate transporter subunit | tRNA-Val | conserved protein | predicted protein | DNA-binding protein, hemimethylated | Qin prophage; predicted protein | predicted S-adenosyl-L-methionine-dependent methyltransferase | cold shock gene | asparagine synthetase A | crotonobetaine reductase subunit II, FAD-binding | antitoxin of the ChpB-ChpS toxin-antitoxin system | serine endoprotease (protease Do), membrane-associated | carbon-phosphorus lyase complex subunit | carbon-phosphorus lyase complex accessory protein | 5S ribosomal RNA of rrnA operon | broad specificity sugar efflux system | sensory histidine kinase in two-component regulatory sytem with UhpA | conserved inner membrane protein | CP4-6 prophage; predicted protein | fused predicted pyruvate-flavodoxin oxidoreductase: conserved protein/conserved protein/FeS binding protein | predicted acetyltransferase |

































































































































































































































































































































































































































































































































































































































